

| ||
| January 3, 2005 [posted] | ||
| PubChem: An Entrez Database of Small Molecules | ||
|
[Editor's Note: This article is a reprint from the Summer/Fall 2004 issue of the NCBI News, an online newsletter from the National Center for Biotechnology Information (NCBI).]
PubChem Substance contains over 800,000 chemical samples imported from 14 public sources including ChemIDplus, the Developmental Therapeutics Program at the National Cancer Institute (NCI), KEGG, NCBI Molecular Modeling Database (MMDB), and the NIST Chemistry WebBook. Chemical entities in PubChem Substance records that have known structures are validated, converted to a standardized form, and imported into PubChem Compound. This standardizing allows NCBI to compute chemical parameters and similarity relationships between compounds. The compounds are grouped into levels of chemical similarity from most general to most specific: same bonding connectivity and any tautomer; same bonding connectivity; same stereochemistry; same isotopes; and same stereochemistry and isotopes. PubChem Compound also indexes these chemicals using 34 fields, many of which represent computed chemical properties such as the number of chiral centers, the number of hydrogen bond donors/acceptors, molecular formula and weight, total formal charge, and octanol-water partition coefficients (XlogP). These groups are provided as Entrez links that allow similar compounds to be retrieved quickly. The third database, PubChem Bioassay, currently includes 173 bioactivity studies from the Developmental Therapeutics Program at NCI, and each of these studies is linked to records in PubChem Substance. The PubChem Bioassay interface allows users to view substances that meet certain activity and/or chemical criteria, and the matching records can either be viewed in PubChem Substance or downloaded in several formats. As part of the Entrez system, the three PubChem databases are linked to several related Entrez databases, including PubMed, Protein, and Structure. PubMed links are derived either from citations provided by submitters or by matching substance names to the MeSH medical thesaurus, which often provide extensive information about the biological activity of a substance. The Protein and Structure links reveal proteins known to interact with a compound and protein structures that contain the compound as a bound ligand. The reverse links also provide new functionalities. Now ligands within structures can be identified instantly by the link to PubChem Compound, as can chemicals described in PubMed abstracts. Consider Gleevec, a potent tyrosine kinase inhibitor used to treat leukemia. In PubChem Substance, the query "gleevec" retrieves one record for Imatinib meslylate from ChemIDplus. Clicking on the SID (substance ID) number or the thumbnail structure loads a Substance Summary showing a view of the structure, other information including chemical properties and synonyms, and inks to PubChem Substance, PubChem Compound, PubMed, and records of identical compounds. This record contains both Imatinib meslylate and methanesulfonic acid; a link to identical compounds leads to substances that also contain the acid. In this case, one additional substance is found that was not retrieved by the query "gleevec", showing how similarity neighboring is able to overcome differing nomenclatures. As part of the standardizing process, substances that have multiple components give rise to several records in PubChem Compound to allow more powerful searching for similar compounds. In the present case, if the Compound Displayed pulldown menu is changed from Standardized to Component1, a different Compound record is shown that contains Imatinib mesylate without the acid, and this compound is linked to seven identical compounds, including itself (Figure 1). Clicking the link to the right of Same Connectivity loads these identical compounds into PubChem Compound, and then choosing Protein Structure from the Display pulldown menu and clicking Display reveals three crystal structures of tyrosine kinase domains containing bound Gleevec. Only one of these structures would have been found by the text query "gleevec" in Entrez Structure, illustrating the advantage of the precomputed chemical similarities provided by PubChem Compound. 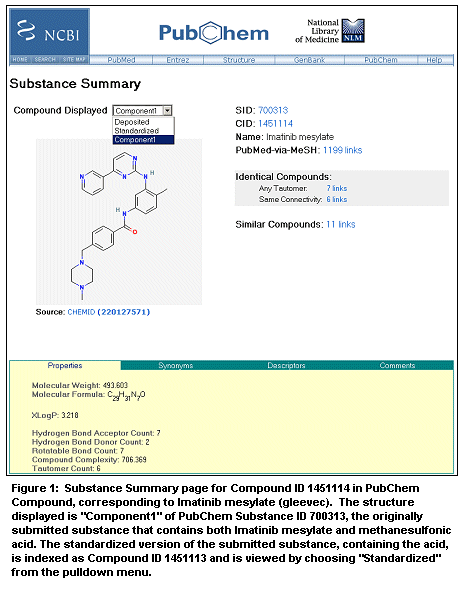
PubChem Bioassay allows one to search for bioactivity. For instance, the query "leukemia AND lc50[tid description]" in PubChem Bioassay retrieves eight growth inhibition assays with measured LC50 values in various leukemia cell lines. Links are then provided to PubChem Substance and PubChem Compound for these chemicals so that they may be further explored. Access PubChem at: http://pubchem.ncbi.nlm.nih.gov.
Questions should be directed to: info@ncbi.nlm.nih.gov
By Eric Sayers Sayers E. PubChem: An Entrez Database of Small Molecules. NLM Tech Bull. 2005 Jan-Feb;(342):e2. | ||

 he NCBI has released three new Entrez databases that link small organic molecules to bioactivity assays, PubMed abstracts, and protein sequences and structures. The new databases compose the
he NCBI has released three new Entrez databases that link small organic molecules to bioactivity assays, PubMed abstracts, and protein sequences and structures. The new databases compose the