Congressional Justification FY 2018
Department of Health and Human Services
National Institutes of Health
National Library of Medicine (NLM)
2018 Congressional Justification (PDF)
FY 2018 Budget
- Organizational Chart
- Appropriation Language
- Amounts Available for Obligation
- Budget Graphs
- Authorizing Legislation
- Appropriations History
- Justification of Budget Request
- Detail of Full-Time Equivalent Employment (FTE)
- Detail of Positions
Organizational Chart

- Office of the Director
Patricia Flatley Brennan, R.N., Ph.D., Director
Betsy L. Humphreys, Deputy Director
Milton Corn, M.D., Deputy Director for Research and Evaluation
Todd D. Danielson, Associate Director for Administrative Management- Division of Extramural Programs
Valerie Florance, Ph.D., Associate Director - Division of Library Operations
Joyce E. B. Backus, Associate Director - Lister Hill National Center for Biomedical Communications
Clem McDonald, M.D., Director - Division of Specialized Information Services
Florence Chang, Acting Associate Director - National Center for Biotechnology Information
David J. Lipman, M.D., Director
- Division of Extramural Programs
Appropriation Language
For carrying out section 301 and title IV of the PHS Act with respect to health information communications, $373,258,000: Provided, That of the amounts available for improvement of information systems, $4,000,000 shall be available until September 30, 2019: Provided further, That in fiscal year 2018, the National Library of Medicine may enter into personal services contracts for the provision of services in facilities owned, operated, or constructed under the jurisdiction of the National Institutes of Health (referred to in this title as "NIH").
Amounts Available for Obligation 1
(Dollars in Thousands)
| Source of Funding | FY 2016 Final | FY 2017 Annualized CR | FY 2018 President's Budget |
|---|---|---|---|
| Appropriation | $394,664 | $394,664 | $373,258 |
| Mandatory Appropriation: (non-add) | |||
| Type 1 Diabetes | (0) | (0) | (0) |
| Other Mandatory financing | (0) | (0) | (0) |
| Rescission | 0 | -751 | 0 |
| Sequestration | 0 | 0 | 0 |
| Zika Intra-NIH Transfer | -546 | 0 | 0 |
| Subtotal, adjusted appropriation | $394,118 | $393,913 | $373,258 |
| OAR HIV/AIDS Transfers | 1,020 | 0 | 0 |
| Subtotal, adjusted budget authority | $395,138 | $393,913 | $373,258 |
| Unobligated balance, start of year | 2,000 | 2,000 | 0 |
| Unobligated balance, end of year | -2,000 | 0 | 0 |
| Subtotal, adjusted budget authority | $395,138 | $395,913 | $373,258 |
| Unobligated balance lapsing | -63 | 0 | 0 |
| Total obligations | $395,074 | $395,913 | $373,258 |
1Excludes the following amounts for reimbursable activities carried out by this account:
FY 2016 - $28,810 FY 2017 - $34,639 FY 2018 - $34,640
Fiscal Year 2018 Budget Graphs
History of Budget Authority and FTEs:
Bar Graph for Funding Levels by Fiscal Year for FY2014 through FY2018
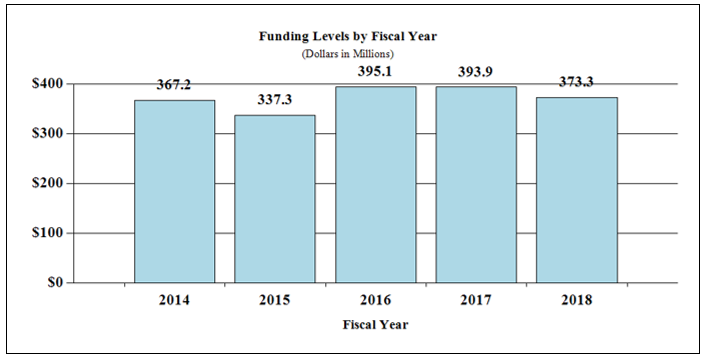
Data for Funding Levels by Fiscal Year for FY2014 through FY2018
| Fiscal Year | Funding (Dollars in Millions) |
|---|---|
| 2014 | $367.2 |
| 2015 | $337.3 |
| 2016 | $395.1 |
| 2017 | $393.9 |
| 2018 | $373.3 |
Bar Graph of FTEs by Fiscal Year for FY2014 through FY2018
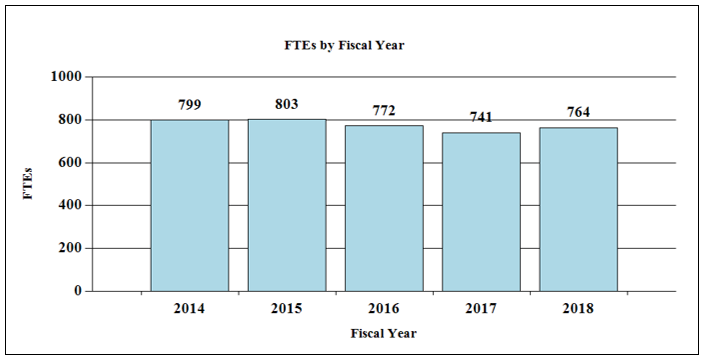
Data for FTEs by Fiscal Year for FY2014 through FY2018
| Fiscal Year | FTEs |
|---|---|
| 2014 | 799 |
| 2015 | 803 |
| 2016 | 772 |
| 2017 | 741 |
| 2018 | 764 |
Authorizing Legislation
| PHS Act/ Other Citation |
U.S. Code Citation |
2017 Amount Authorized |
FY 2017 Annualized CR | 2017 Amount Authorized |
FY 2018 President's Budget | |
|---|---|---|---|---|---|---|
| Research and Investigation | Section 301 | 42§241 | Indefinite | $393,913,000 | Indefinite | $373,258,000 |
| National Library of Medicine | Section 401(a) | 42§281 | Indefinite | Indefinite | ||
| Total, Budget Authority | $393,913,000 | $373,258,000 | ||||
Appropriations History
| Fiscal Year | Budget Estimate to Congress | House Allowance | Senate Allowance | Appropriation |
|---|---|---|---|---|
| 2008 | $312,562,000 | $325,484,000 | $327,817,000 | $326,669,000 |
| Rescission | $5,707,000 | |||
| 2009 | $323,046,000 | $331,847,000 | $329,996,000 | $330,771,000 |
| Rescission | $0 | |||
| Supplemental | $1,705,000 | |||
| 2010 | $334,347,000 | $342,585,000 | $336,417,000 | $339,716,000 |
| Rescission | $0 | |||
| 2011 | $364,802,000 | $364,254,000 | $339,716,000 | |
| Rescission | $2,982,909 | |||
| 2012 | $387,153,000 | $387,153,000 | $358,979,000 | $338,278,000 |
| Rescission | $639,345 | |||
| 2013 | $372,651,000 | $381,981,000 | $337,638,655 | |
| Rescission | $675,277 | |||
| Sequestration | ($16,947,139) | |||
| 2014 | $382,252,000 | $387,912,000 | $327,723,000 | |
| Rescission | $0 | |||
| 2015 | $372,851,000 | $336,939,000 | ||
| Rescission | $0 | |||
| 2016 | $394,090,000 | $341,119,000 | $402,251,000 | $394,664,000 |
| Rescission | $0 | |||
| 20171 | $395,684,000 | $407,086,000 | $412,097,000 | $394,664,000 |
| Rescission | $0 | |||
| 2018 | $373,258,000 |
1 Budget Estimate to Congress includes mandatory financing.
Justification of Budget Request
Authorizing Legislation: Section 301 and title IV of the Public Health Service Act, as amended.
Budget Authority (BA):
| FY 2016 Actual |
FY 2017 Annualized CR |
FY 2018 President’s Budget |
FY 2018 +/- FY 2017 |
|
|---|---|---|---|---|
| BA | $395,137,766 | $393,913,000 | $373,258,000 | -$20,655,000 |
| FTE | 772 | 741 | 764 | +23 |
Program funds are allocated as follows: Competitive Grants/Cooperative Agreements; Contracts; Direct Federal/Intramural and Other.
Director's Overview
The National Library of Medicine (NLM), the world’s largest medical library and a leader in biomedical data science, acquires, organizes, and delivers up-to-date biomedical information across the United States and around the globe. Through its information systems, cutting-edge data science and informatics research, extensive training programs, and many partnerships, NLM plays an essential role in catalyzing basic science; making research results available for translation into new treatments, products, and practices; providing useful decision support for health professionals and patients; and supporting disaster and emergency preparedness and response. NLM coordinates a 6,400 member National Network of Libraries of Medicine (NN/LM) that improves access to health information in communities nationwide, with an emphasis on reaching populations with health disparities.
Millions of scientists, health professionals, and members of the public use NLM’s electronic information sources billions of times each year. The range of information that NLM assembles and disseminates is enormous, including genetic, genomic, biochemical, and toxicological data; clinical and scientific images; published research results; historical archives; information about clinical trials and FDA-regulated drugs and devices; scientific and health data standards; informatics tools for system developers; and health information for the public. NLM dispatches an average of more than 6,500 million bits of data per second, making it one of the Federal government’s largest providers of digital content. Anyone can search or download information directly from an NLM web site, find it via an Internet search engine, or use an “app” that provides value-added access to NLM data. Thousands of commercial and non-profit system developers regularly use the applications programming interfaces (APIs) that NLM provides to fuel private sector innovation and to embed access to NLM information services within electronic health records (EHRs). NLM provides mission-critical services to researchers, hospitals, and industry.
Priorities for FY 2018
Fundamental Science. High-quality, cost-effective science evaluates and builds upon evidence in published papers, makes effective use of existing scientific data, applies rigorous methods, publishes complete descriptions of methods and results, and shares new data that, in turn, promote new science and new discoveries. NLM’s rapidly growing and richly linked databases and systems promote scientific breakthroughs by playing an essential role in all phases of this process. Every day, NLM receives ten terabytes of new data and publications, adds value by enhancing quality and consistency and integrating them with other information in NLM databases, and responds to millions of inquiries from individuals and computer systems by serving up more than 75 terabytes of information, including three million published journal articles.
Scientists around the world analyze data and information using software, algorithms, and methods arising from basic research in computational biology and biomedical informatics conducted or funded by NLM. For example, tools developed by NLM researchers are used to analyze high throughput genomic data to identify promising drug targets, to mine journal articles and EHRs to discover adverse drug reactions, and to detect transplant rejection earlier so that interventions to help clinical research participants can begin more quickly. Notably, a major breakthrough in gene editing technology, CRISPR-Cas, had a foundation in the work on bacterial evolutionary genomics by an intramural research group at NLM.
Treatments and Cures. The acquisition and use of new knowledge about the health effects of an individual’s genetic makeup, environmental exposures, lifestyle choices and constraints, and health care interventions are necessary prerequisites to precision medicine. NLM supports both the discovery and the direct clinical application of such knowledge. The Database of Genotypes and Phenotypes (dbGaP) includes the results of approximately 700 studies of the interaction between genetic makeup and observable traits (high cholesterol, for example) associated with certain diseases. The ClinVar database makes knowledge about the clinical significance of scientifically validated genetic variations available to clinicians. ClinicalTrials.gov includes an increasing number of studies that identify the genetic variations associated with different outcomes to provide confirming evidence for precision medicine.
The NIH All of UsSM Research Program aims to capitalize on relatively inexpensive genome sequencing technology, growing availability of Electronic Health Records (EHRs) – many employing standard clinical vocabularies developed, supported, and disseminated by NLM, and participants’ ability to provide data directly via smart phones, sensors, and other mobile devices. Extramural scientists trained and funded by NLM are leaders in development of methods for extracting – and quantifying the reliability of – data analytics from large quantities of disparate and incomplete EHR data.
Health Promotion and Disease Prevention. Inexpensive genome sequence technology also plays a role in detecting and preventing the spread of outbreaks of foodborne illness. In a partnership with the Centers for Disease Control and Prevention (CDC), the Food and Drug Administration (FDA), the Department of Agriculture, Public Health England, and state and regional laboratories, NLM’s National Center for Biotechnology Information (NCBI) receives whole genome sequences of pathogens collected from patients, food, and food production facilities; rapidly processes and compares them to others in NLM databases to identify closely related sequences; and returns the results within 24 hours, enabling faster detection and response to outbreaks. In addition, NLM’s pathogen detection system and its new set of annotated records for DNA sequences that encode proteins contributing to resistance to various antibiotics will make essential contributions to national efforts to combat antibiotic resistant bacteria.
These are important advances, but health promotion and disease prevention are critically dependent on individual behavior and community action. NLM makes extensive free, high quality health information available to individuals and communities as they confront health problems and emergencies. Through the NN/LM and partnerships with minority-serving institutions, tribal organizations, public health departments, schools, churches, and others, NLM supports projects to increase awareness of and access to health information in communities across the nation.
Enhancing Stewardship. NLM’s PubMed, PubMed Central, and ClinicalTrials.gov are important tools that help NIH to measure the output of research investments and assess and comment on the rigor and reproducibility of research results. NLM databases also enable NIH and other research funders to monitor compliance with requirements for public access to publications and clinical trial results from their funded studies. NLM expects a significant increase in results submissions to ClinicalTrials.gov due to a recent final regulation affecting many trials of FDA-regulated drugs and devices and a new NIH policy that applies to all NIH-funded trials. Maintaining and enhancing robust 24/7 availability of this and other critical information services are enduring stewardship priorities for NLM, in the face of rapid growth in content submissions, system use, and cybersecurity threats. NLM’s extensive training programs will continue to be an important source of informatics and data science researchers and a mechanism for promoting research rigor and reproducibility in these fields.
Overall Budget Policy:
The FY 2018 President’s Budget request is $373.258 million, a decrease of $20.655 million below the FY 2017 Annualized Continuing Resolution (CR) level. Funds are included to maintain NLM’s most heavily used services that provide access to published biomedical literature and consumer health information. At this level, support for some NLM databases and services will be scaled back or discontinued. NLM will support key intramural data science activities. These include processing and organizing genomic data resulting from NIH-wide investments and from whole genome sequencing of pathogens to support outbreak detection and response by CDC, FDA, and USDA; managing and enhancing ClinicalTrials.gov, the world’s largest clinical trials registration and summary results reporting system; updating and disseminating clinical terminology standards required for U.S. health data interoperability; and conducting research in data science methods, standards, and tools and in advanced integration and analysis of genomic data, clinical research data, and observational health data. NLM will continue to support the National Network of Libraries of Medicine and its role in improving U.S.-wide access and use of health information and scientific data in communities across the nation. NLM will streamline, consolidate, or scale back other outreach programs designed to promote access and training in the effective use of NLM resources for scientists, clinicians, patients, and the general public.
Program Descriptions and Accomplishments
Intramural Programs
NLM’s intramural programs encompass the development, maintenance, and delivery of information services and stewardship of the published record of biomedical knowledge, as well as intramural research and training. In NLM’s case, intramural research and training is focused on computational biology, biomedical informatics, data science, and information science.
Delivering Reliable, High Quality Biomedical and Health Information Services: In FY 2016, NLM greatly expanded the quantity and range of high quality information readily available to scientists, health professionals, and the general public. Advances included:
- indexing of approximately 806,000 new journal articles for PubMed/MEDLINE, NLM’s most heavily used database, which contains records for more than 26 million articles in biomedical and life sciences journals and delivers information to more than four million users per day;
- growth in the PubMed Central (PMC) digital archive, which now includes more than four million full-text research articles, including those produced by researchers funded by NIH and ten other government agencies;
- expansion of ClinicalTrials.gov, the world’s largest clinical trials registry, which now includes more than 228,000 registered studies and summary results for more than 22,000 studies, including many not elsewhere published;
- enhancement of Genetics Home Reference (GHR), which provides consumer-level information about 2,500 genetic conditions, genes, gene families, and chromosomes to an average of 1.9 million visitors per month;
- more than 20 percent growth in dbGaP, which connects individual-level genomic data with individual-level clinical information and now contains almost 700 studies involving more than 1.2 million people;
- continued growth of PubChem, an archive of chemical and biological data on small molecules; PubChem contains information on more than 90 million unique chemical structures and more than 1.2 million bioassays;
- expansion of the RefSeq database of integrated, non-redundant, well-annotated reference sequences, which are essential to identifying and documenting genetic variations that affect human health, to over 20 million genomic records, a 35 percent increase in FY 2016, and more than 70 million protein records from over 62,000 organisms;
- release of MedPix®, a fully web-enabled cross-platform database, integrating more than 53,000 clinical images and related text originally collected at the Uniformed Services University of the Health Sciences, with a primary target audience of physicians, nurses, allied health professionals, and students; and
- launch of AccessGUDID, a database that already provides access to unique identifiers and registration information submitted to FDA for more than one million medical devices, including implantables, to support improvements in care and patient safety.
NLM also continued to expand access to its rare and unique historical collections through digitization partnerships with outside organizations. In FY 2016, more than 1,600 printed historical books and 37,000 images were digitized and added to NLM’s Digital Collections, a free online archive of biomedical books and videos. These collections are heavily used by scholars, the media, and the general public.
Program Portrait: Enhancing the Interoperability of Electronic Health Records (EHRs)
FY 2017 Level: $18.3 million
FY 2018 Level: $17.3 million
Change: -$1.0 million
In close collaboration with the Office of the National Coordinator for Health Information Technology within HHS and with support from Centers for Medicare and Medicaid Services (CMS), the Veterans Health Administration, and FDA, NLM develops, funds, and disseminates the clinical terminologies designated as U.S. standards for meaningful use of EHRs and health information exchange. The goal is to ensure that EHR data created in one system can be transmitted, interpreted, and aggregated appropriately in other systems to support health care, public health, and research. NLM produces a range of tools that help EHR developers and users to implement these standards and makes them available in multiple formats, including via application programming interfaces (APIs). NLM’s technical and financial support enables clinical terminology standards to be updated regularly to reflect new drugs, tests, devices, and changes in medical knowledge and health practice. In FY 2016, for example, NLM worked quickly with CDC and the standards community to develop terminology needed to support response to the evolving Zika virus outbreak. Importantly, NLM’s financial support also allows key standards to be used free-of-charge in U.S. health care, public health, biomedical research, and product development.
Inclusion of standard terminology in EHRs makes it easier to use information in a patient’s record to retrieve relevant electronic knowledge and provide it to the patient. In FY 2016, NLM’s MedlinePlus Connect service, which enables vendors to connect EHR products directly to NLM’s high quality information relevant to a patient’s problems, medications, and test results, fielded more than 71 million requests for information. Standardized terminology also facilitates other forms of automated clinical decision support and quality measurement. NLM’s Value Set Authority Center (VSAC) enables people to collaborate on defining sets of standard terms to be used, for example, to identify currently marketed prescription opioid drugs or to identify diabetic patients in a clinician’s practice who haven’t received a foot check-up. VSAC is the authoritative source of value sets that are part of CMS-required quality measures. It is also a key resource in efforts to facilitate use of standard clinical terminology and common data elements in NIH-funded clinical research. NLM’s Unified Medical Language System (UMLS) resources connect standard clinical terminologies to billing codes and more than 120 other important biomedical vocabularies, such as those used in information retrieval and gene annotation. By linking many different names for the same concepts and by providing associated natural language processing tools, UMLS resources help computer programs to interpret biomedical text and health data correctly in NIH-funded research, in commercial product development, and in many electronic information services, including those produced by NLM.
NLM continued efforts to redesign its web interfaces to use “responsive designs” that automatically adjust the information display to match the size of the device, such as smart phones. In FY 2016, NLM released new responsive design versions of four heavily used resources.
Promoting Public Awareness and Access to Information: While NLM services receive tremendous use and are effectively publicized via social media and other channels, NLM also uses other means, like church meetings and health fairs, to highlight its services to diverse populations in ways most acceptable to them. NLM works with the NN/LM; formal outreach partnerships, including the Partners in Information Access for the Public Health Workforce and the Environmental Health Information Outreach Partnership with Historically Black Colleges and Universities, tribal colleges, and other minority serving institutions; and more informal community partnerships to extend its reach into diverse communities across the nation. In FY 2016, NLM funded more than 200 outreach projects across the country to enhance awareness and access to health information, including in disaster and emergency situations, and to address health literacy issues.
NLM also uses exhibitions, the media, and new technologies in its efforts to reach underserved populations and to promote interest among young people in science, medicine, and technology. NLM continues to expand its successful traveling exhibitions program as another means to enhance access to its services and promote interest in science and medicine in communities across the country. In FY 2016, public-private partnerships enabled NLM traveling exhibitions to appear in 161 institutions in 144 towns and cities in 42 states. Examples include: Physician Assistants: Collaboration and Care; Native Voices: Native Peoples’ Concepts of Health and Illness; and Surviving and Thriving: AIDS, Politics, and Culture.
Program Portrait: National Network of Libraries of Medicine (NN/LM)
FY 2017 Level: $12.0 million
FY 2018 Level: $12.0 million
Change: $0.0 million
The 6,400 member institutions of the NN/LM are valued partners in ensuring that health information, including NLM’s many services, is available to scientists, health professionals, and the public. NN/LM is coordinated by eight Regional Medical Libraries and is comprised of academic health sciences libraries, hospital libraries, public libraries, and community-based organizations.
Based on stakeholder feedback, in FY 2016, NLM changed the mechanism used to support the Regional Medical Libraries from contracts to cooperative agreements (grants) to provide greater flexibility to meet changing priorities and user needs and behaviors. Awards were made to health sciences libraries at the University of Massachusetts; the University of Pittsburgh; the University of Maryland; the University of Iowa; the University of Utah; the University of North Texas; the University of California, Los Angeles; and the University of Washington. Under these cooperative agreements, the NN/LM is governed by a National Network Steering Committee (NNSC), which is working together to ensure better coordination among regions and less duplication of effort for services that can be delivered nationwide by one library to all regions. The NN/LM will continue to play a pivotal role in outreach by exhibiting and demonstrating NLM's products and services at national, regional, and state health professional and consumer oriented meetings; coordinating efforts to improve access to electronic publications for the public health workforce; improving awareness and access to high quality health information for the general public; addressing health literacy and health disparities; and supporting recruitment and participant engagement in the NIH All of Us Precision Medicine initiative
The NN/LM has an excellent track record of providing access to health information in disasters and emergencies and will continue to serve as the backbone of NLM's strategy to promote more effective use of libraries and librarians in local, state, and national disaster preparedness and response efforts. NN/LM also plays an important role in NLM efforts to increase the capacity of research libraries and librarians to support data science and improve institutional capacity in biomedical big data management and analysis.
As a collaborative venture with NIH Institutes and Centers (ICs) and other partners, NLM produces the NIH MedlinePlus magazine, and its Spanish counterpart, NIH Salud. The magazine, which is also available online in Spanish and English, is distributed without charge to 70,000 individual subscribers, well as to doctors’ offices, health science libraries, Congress, the media, federally supported community health centers, select hospital emergency and waiting rooms, and other locations where the public receives health services nationwide. In FY 2016, NLM and NIH partnered with the National Hispanic Medical Association, the American Diabetes Association, and the Peripheral Arterial Disease Coalition, among others, to extend the distribution of the magazine to the audiences they serve.
Developing Advanced Information Systems, Standards, and Research Tools: NLM’s advanced information services have long benefitted from its intramural research and development (R&D) programs, which have led to major advances in the ways high volume information and data are collected, structured, standardized, mined, and delivered. The Library has two organizations that conduct advanced R&D on different aspects of biomedical informatics – the Lister Hill National Center for Biomedical Communications (LHC) and NCBI.
LHC, established by joint resolution of Congress in 1968, conducts and supports R&D in such areas as the development and dissemination of health data standards; the capture, processing, dissemination, and use of high quality imaging data; medical language processing; high-speed access to biomedical information; advanced technology for emergency and disaster management; and analysis of large databases of clinical and administrative data to determine their usefulness in predicting patient outcomes and in validating findings from relatively small prospective clinical research studies. In FY 2016, for example, researchers employed the Massachusetts Institute of Technology’s large MIMIC-III database of de-identified intensive care data to advance R&D in text retrieval techniques to enhance computer-assisted clinical decision support based on free-text clinical notes.
LHC’s ongoing research involving language resources and innovative algorithms and tools, including the UMLS, MetaMap, Medical Text Indexer (MTI), and SemRep, serves to advance research in natural language understanding and biomedical text mining. This research has been applied to indexing, information retrieval, question answering, and literature-based discovery to assist NLM’s high volume data creation and service operations, to help other NIH components to identify and summarize new knowledge useful in updating clinical guidelines, and to add standard terminology and codes to clinical data to enhance their research value.
NLM has continued to expand and enhance its extensive collections of open imaging data and open software, including the heavily used Insight TookKit for three-dimensional (3D) analysis of biomedical imaging data (the ITK toolkit). Algorithms and software received through the FY 2016 Pill Image Recognition Challenge were evaluated to rank how well consumer images of prescription pills match reference images of pills in the authoritative NLM RxIMAGE database. LHC is a major contributor to the NIH 3D Print Exchange, including collections and models useful to radiologists, surgeons, and prosthetics and robotics experts. In FY 2016, NLM began to develop an open source annotated library of anatomical models derived from medical imaging data that are high quality, accurate, and optimized for use with interactive development, 3D printing, animation, and virtual reality.
NLM leverages its imaging research and analysis expertise to address critical clinical and global health needs, in collaboration with the National Cancer Institute (NCI) and the National Institute of Allergy and Infectious Disease (NIAID), as well as other biomedical research and healthcare delivery organizations. In FY 2016, LHC scientists made chest X-ray datasets publicly available to enable scientific research. The datasets have been downloaded by more than 200 academic and industrial researchers worldwide and were used to conduct R&D in computer-assisted radiology image analysis and improve the performance of algorithms that detect probable tuberculosis in chest X-rays. To improve malaria diagnosis and management in the field, LHC researchers won an HHS Ventures Innovation award that supported the development of a MalariaScreener smartphone application to automatically detect and count parasitic cells in a blood smear slide, in collaboration with NIAID and the Oxford-Mahidol Center in Thailand. In FY 2016, field-testing of the mobile app was started in Bangladesh in collaboration with the Bangladesh Malaria Control Program.
NCBI, established by law in 1988, conducts R&D on the representation, integration, and retrieval of molecular biology data and biomedical literature, in addition to providing an integrated genomic information resource consisting of more than 40 databases for biomedical researchers at NIH and around the world. NCBI’s development of large-scale data integration techniques with advanced information systems is key to its expanding ability to support the accelerated pace of research made possible by new technologies such as next-generation DNA sequencing, microarrays, and small molecule screening. GenBank at NCBI, in collaboration with partners in the United Kingdom and Japan, is the world’s largest annotated collection of publicly available DNA sequences. GenBank contains 196 million sequences from more than 370,000 different species. NCBI’s web services for access to these data provide the information and analytic tools for researchers to accelerate the rate of genomic discovery and facilitate the translation of basic science advances into new diagnostics and treatments.
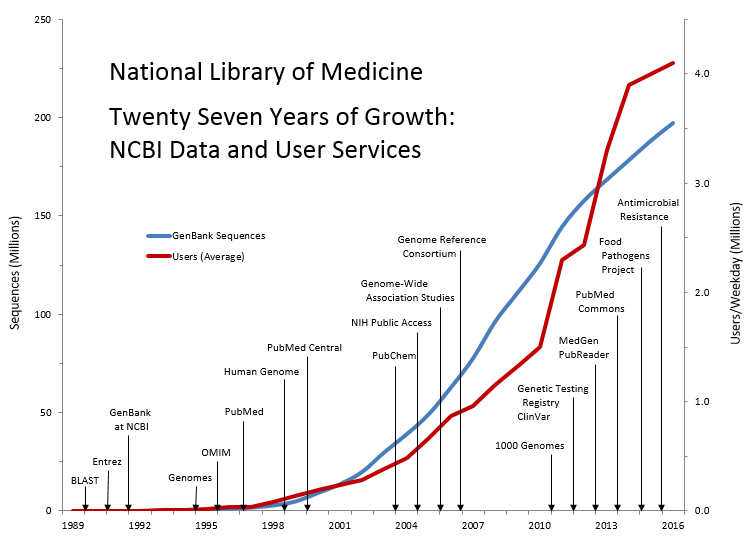
Data for Twenty-Seven Years of Growth: NCBI Data and User Services
| Year | Product Release | GenBank Sequences | Users (Average) |
|---|---|---|---|
| 1989 | 30,010 | 0 | |
| 1990 | BLAST | 40,295 | 0 |
| 1991 | Entrez | 58,952 | 0 |
| 1992 | GenBank at NCBI | 87,846 | 0 |
| 1993 | 143,492 | 4,500 | |
| 1994 | 215,273 | 9,000 | |
| 1995 | Genomes | 555,694 | 19,000 |
| 1996 | OMIM | 1,021,211 | 36,000 |
| 1997 | PubMed | 1,765,847 | 39,268 |
| 1998 | 2,837,897 | 84,256 | |
| 1999 | Human Genome | 4,864,570 | 138,139 |
| 2000 | PubMed Central | 9,102,634 | 190,485 |
| 2001 | 13,602,262 | 233,977 | |
| 2002 | 19,808,101 | 283,590 | |
| 2003 | 29,819,397 | 380,104 | |
| 2004 | PubChem | 38,941,263 | 485,029 |
| 2005 | NIH Public Access | 49,152,445 | 666,917 |
| 2006 | Genome-Wide Association Studies | 62,765,195 | 864,586 |
| 2007 | Genome Reference Consortium | 77,632,813 | 958,584 |
| 2008 | 96,400,790 | 1,152,700 | |
| 2009 | 110,946,879 | 1,328,143 | |
| 2010 | 1000 Genomes | 125,764,384 | 1,501,335 |
| 2011 | 144,458,648 | 2,300,000 | |
| 2012 | Genetic Testing Registry and ClinVar | 157,889,737 | 2,430,000 |
| 2013 | MedGen and PubReader | 168,335,396 | 3,300,000 |
| 2014 | Pubmed Commons | 178,322,253 | 3,900,000 |
| 2015 | Food Pathogens Project | 188,372,017 | 4,000,000 |
| 2016 | Antimicrobial Resistance | 197,390,691 | 4,100,000 |
As part of the President’s National Action Plan for Combating Antibiotic-Resistant Bacteria, NCBI collaborates with FDA, the CDC, the Department of Agriculture, and other groups to maintain a database of whole genome sequencing (WGS) data for antibiotic-resistant bacteria along with tools to facilitate analyses of such data. The database provides an important resource for surveillance and research into the mechanisms underlying the emergence of antibacterial resistance. This program builds upon a successful collaborative project among these same agencies to use WGS to more quickly and accurately identify and investigate outbreaks of disease caused by foodborne pathogens.
Extramural Programs
NLM funds extramural research, resource, and workforce development programs that provide important foundations for the field of biomedical informatics and biomedical data science, which brings the methods and concepts of computational, informational, quantitative, social/behavioral and engineering sciences to bear on problems related to basic biomedical/behavioral research, health care, public health, and consumer use of health-related information. Research focuses on the development and testing of approaches for acquiring, integrating, managing, mining, analyzing, and presenting biomedical data, information, and knowledge across a spectrum of sizes and data types. In addition to grants for basic and applied research, predoctoral and postdoctoral training, and career development, NLM sponsors several unique resource grant programs that support biomedical knowledge resource developments. To accomplish its extramural goals in FY 2018, NLM will offer grants in four general categories: research project grants and supplements; training/fellowship/career support; information resource awards; and small business grants. NLM will also continue to provide management oversight for a selection of grants for NIH pioneer and early innovation awards and data science research training and digital curation awards, funded by the NIH Common Fund
Informatics Workforce and Resources for Biomedicine and Health: Many of today’s informatics researchers and health information technology leaders are graduates of NLM-funded university-based training programs in biomedical informatics. NLM’s 14 active university-based programs train more than 200 individuals each year. NLM held a new competition for university-based training programs in FY 2016, expanding its emphasis on data science and support for clinician researchers with new awards anticipated starting in the summer of 2017. Two career transition programs are offered to NLM’s trainees and others ready to launch their informatics research careers. Taken together, NLM’s commitment to training and career transition in FY 2016 represented nearly 33 percent of NLM’s extramural grants budget. In FY 2018, NLM will continue to support research training and career transition through institutional training programs and grants to individuals.
NLM has three unique resource grant programs offered by no other Federal agency. Grants for Scholarly Works in Biomedicine and Health support researchers in the history and philosophy of medicine, biomedical science, and bioethics. NLM Administrative Supplements for Informationist Services provide supplemental funds to existing NIH research grantees who want to add an information specialist to their research team. These awards foster better management of biomedical research data, making the work of these awardees foundational for the application of big data to improve health. Eleven two-year awards, five of them partially or fully funded by other ICs, continued in FY 2016. Some three-year informationist supplement awards will be made in FY 2017.
The third unique program, NLM Information Resources to Reduce Health Disparities, has a unique focus on development of information resources; four three-year awards supporting development of information resources tailored to needs of Alaska Native, Navajo, and other underserved populations continued in FY 2016. Resources like these support long-term goals of the All of Us Research Program to engage a large cohort of active participants. In FY 2017, NLM expects to award up to five new three-year information resource projects.
Biomedical Informatics Research: NLM research project grants (RPGs) have supported pioneering research and development in computational intelligence for medicine, clinical decision support, protection of privacy in electronic medical records, secondary use of routine clinical data for research purposes, regional health data integration, consumer use of personal health information, health applications of advanced telecommunications networks, and automated bio-surveillance for public health. At the core, these projects advance the science of biomedical informatics by applying concepts from computer, information, quantitative, social/behavioral and engineering sciences to problems in medicine, public health, consumer health, and biological sciences. Complementing and building upon the BD2K and informatics-related initiatives at other ICs, NLM research grant programs continue to support innovation in both basic and applied research ranging from multi-site research collaborations to small proof-of-concept projects. In all research supported by NLM, needs of the intended user base must be taken into account, whether they are researchers, students, clinicians, health administrators, patients, or consumers. NLM funds investigator-initiated projects, as well as projects from focused funding announcements that target areas important to NLM’s mission.
In FY 2016, NLM issued 16 new RPGs including three exploratory/developmental awards and one Academic Research Enhancement Award. Among the newly funded research awards are several that support improved patient engagement, including use of digital images to diagnose patients at risk of diabetic retinopathy and testing the ability of consumer health technology to foster improved communication between diabetics and their clinician caregivers. Three new awards in translational bioinformatics focus on modeling heterogeneity, metabolism, and drug uptake in cells and cellular processes. Several of the new awards address big data topics, including research into improved causal inference methods and exploratory techniques for reproducible research using observational data. Three of the new awardees are developing approaches to use data in EHRs to improve health care outcomes. In addition to new and continuing awards, NLM made one supplemental award to an existing NLM research grant. In FY 2018, NLM expects to award new RPGs with increased focus on patient engagement and understandable health information to support decisions by clinicians and patients.
NLM sets aside funds to support small business innovation and research and technology transfer (SBIR/STTR). In FY 2016, NLM’s allocation of funds for SBIR/STTR increased to more than $1 million for the first time; NLM awarded four new projects, three of which center on improvements to management and use of EHR data and one on parallel processing of biomedical big data. NLM will meet the required set-aside and expects to award three or four SBIR/STTR awards in FY 2017.
Program Portrait: Research in Biomedical Informatics and Data Science
FY 2017 Level: $26.2 million
FY 2018 Level: $26.5 million
Change: +$0.3 million
For more than 30 years, NLM’s Extramural Programs Division has been a principal source of NIH support for research in basic and applied biomedical informatics and data science. NLM’s portfolio of funded research has spanned artificial intelligence, computational biology, physician decision support, public health surveillance, and visualization and discovery mining in digital data sets. NLM’s scope of research interests is wide, encompassing data science and informatics research areas of high importance to NIH and society at large for audiences ranging from clinicians to scientists to consumers and patients.
For example, NLM has recently funded grants that address important data science issues, including:
- accounting for bias, missing data, and other inconsistencies when using data from EHRs in research;
- enhancing the value of existing data from intensive care units for real-time decision making by understanding the timing of gradual state changes and validating causation assertions made using uncertain data;
- testing methods used to extract information from published articles about relationships among drugs, genes, and phenotypes to see if they are also effective on text in EHRs;
- developing tools to help researchers understand the heterogeneity of single cancer cells in a tumor by allowing multiple genomic assays of a single cell, to assist in identifying effective cancer therapies; and
- analyzing formal and informal data sources, e.g., social media, to determine their value in predicting outbreaks of foodborne illness at local and national scale.
NLM will continue to fund research on a wide range of biomedical informatics and data science questions. In addition, NLM will expand its research into analytic approaches and visualization tools that help patients, consumers, and clinicians participate actively in precision medicine research.
Research Management and Support (RMS)
RMS activities provide administrative, budgetary, logistical, and scientific support for basic library services, intramural research programs, and the review, award, and monitoring of research grants and training awards. RMS functions also include strategic planning, coordination, and evaluation of NLM’s programs, regulatory compliance, policy development, and international coordination and liaison with other Federal agencies, Congress, and the public. These activities are conducted by the NLM Director and immediate staff, as well as NLM personnel from the Office of Extramural Programs, the Office of Administrative Management, the Office of Health Information Programs Development, and the Office of Communications and Public Liaison. The NLM Board of Regents is currently overseeing the development of a new NLM Strategic Plan.
Detail of Full-Time Equivalent Employment (FTEs)
| OFFICE/DIVISION | FY 2016 Final | FY 2017 Annualized CR | FY 2018 President's Budget | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Civilian | Military | Total | Civilian | Military | Total | Civilian | Military | Total | |
| Division of Extramural Programs | |||||||||
| Direct: | 21 | - | 21 | 21 | - | 21 | 21 | - | 21 |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 21 | - | 21 | 21 | - | 21 | 21 | - | 21 |
| Division of Library Operations | |||||||||
| Direct: | 282 | - | 282 | 267 | - | 267 | 271 | - | 271 |
| Division of Library Operations | |||||||||
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Division of Library Operations | |||||||||
| Total: | 282 | - | 282 | 267 | - | 267 | 271 | - | 271 |
| Division of Specialized Information Services | |||||||||
| Direct: | 39 | - | 39 | 36 | - | 36 | 37 | - | 37 |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 39 | - | 39 | 36 | - | 36 | 37 | - | 37 |
| Lister Hill National Center for Biomedical Communications | |||||||||
| Direct: | 60 | - | 60 | 52 | - | 52 | 61 | - | 61 |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 60 | - | 60 | 52 | - | 52 | 61 | - | 61 |
| National Center for Biotechnology Information | |||||||||
| Direct: | 273 | 1 | 274 | 265 | 1 | 266 | 274 | 1 | 275 |
| Reimbursable: | 22 | - | 22 | 22 | - | 22 | 22 | - | 22 |
| Total: | 295 | 1 | 296 | 287 | 1 | 288 | 296 | 1 | 297 |
| Office of the Director/Administration | |||||||||
| Direct: | 59 | - | 59 | 62 | - | 62 | 62 | - | 62 |
| Reimbursable: | 15 | - | 15 | 15 | - | 15 | 15 | - | 15 |
| Total: | 74 | - | 74 | 77 | - | 77 | 77 | - | 77 |
| Total | 771 | 1 | 772 | 740 | 1 | 741 | 763 | 1 | 764 |
| Includes FTEs whose payroll obligations are supported by the NIH Common Fund. | |||||||||
| FTEs supported by funds from Cooperative Research and Development Agreements. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FISCAL YEAR | Average GS Grade | ||||||||
| 2014 | 11.2 | ||||||||
| 2015 | 11.4 | ||||||||
| 2016 | 11.5 | ||||||||
| 2017 | 11.5 | ||||||||
| 2018 | 11.5 | ||||||||
Detail of Positions 1
| GRADE | FY 2015 Actual | FY 2016 Enacted | FY 2017 President's Budget |
|---|---|---|---|
| Total, ES Positions | 5 | 5 | 5 |
| Total, ES Salary | 883,612 | 897,131 | 914,804 |
| GM/GS-15 | 29 | 29 | 29 |
| GM/GS-14 | 49 | 49 | 49 |
| GM/GS-13 | 134 | 103 | 126 |
| GS-12 | 129 | 129 | 129 |
| GS-11 | 37 | 37 | 37 |
| GS-10 | 0 | 0 | 0 |
| GS-9 | 16 | 16 | 16 |
| GS-8 | 42 | 42 | 42 |
| GS-7 | 12 | 12 | 12 |
| GS-6 | 1 | 1 | 1 |
| GS-5 | 4 | 4 | 4 |
| GS-4 | 2 | 2 | 2 |
| GS-3 | 7 | 7 | 7 |
| GS-2 | 3 | 3 | 3 |
| GS-1 | 8 | 8 | 8 |
| Subtotal | 473 | 442 | 465 |
| Grades established by Act of July 1, 1944 (42 U.S.C. 207) | 0 | 0 | 0 |
| Assistant Surgeon General | 0 | 0 | 0 |
| Director Grade | 0 | 0 | 0 |
| Senior Grade | 0 | 0 | 0 |
| Full Grade | 0 | 0 | 0 |
| Senior Assistant Grade | 0 | 0 | 0 |
| Assistant Grade | 1 | 1 | 1 |
| Subtotal | 1 | 1 | 1 |
| Ungraded | 290 | 290 | 290 |
| Total permanent positions | 449 | 418 | 441 |
| Total positions, end of year | 781 | 750 | 773 |
| Total full-time equivalent (FTE) employment, end of year | 772 | 741 | 764 |
| Average ES salary | 176,722 | 179,426 | 182,961 |
| Average GM/GS grade | 11.5 | 11.5 | 11.5 |
| Average GM/GS salary | 97,013 | 98,497 | 100,437 |
1 Includes FTEs whose payroll obligations are supported by the NIH Common Fund.
Last Reviewed: February 10, 2026

