Congressional Justification FY 2020
Department of Health and Human Services
National Institutes of Health
National Library of Medicine (NLM)
2020 Congressional Justification (PDF)
FY 2020 Budget
- Organization Chart
- Appropriation Language
- Amounts Available for Obligation
- Budget Mechanism Table
- Major Changes in Budget Request
- Summary of Changes
- Budget Graphs
- Budget Authority by Activity
- Authorizing Legislation
- Appropriations History
- Justification of Budget Request
- Budget Authority by Object Class
- Salaries and Expenses
- Detail of Full-Time Equivalent Employment (FTE)
- Detail of Positions
Organizational Chart
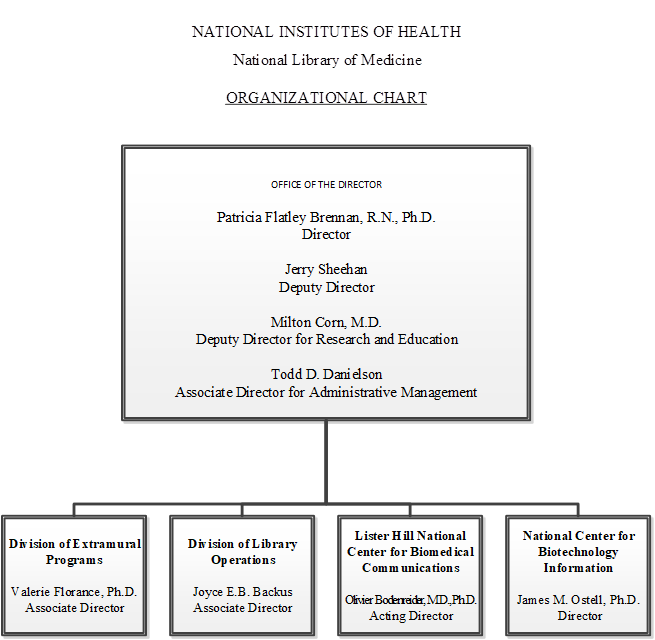
- Office of the Director
Patricia Flatley Brennan, R.N., Ph.D., Director
Jerry Sheehan, Deputy Director
Milton Corn, M.D., Deputy Director for Research and Evaluation
Todd D. Danielson, Associate Director for Administrative Management- Division of Extramural Programs
Valerie Florance, Ph.D., Director - Division of Library Operations
Joyce E. B. Backus, Associate Director - Lister Hill National Center for Biomedical Communications
Olivier Bodenreider, M.D., PhD., Acting Director - National Center for Biotechnology Information
James M. Ostell, Ph.D., Director
- Division of Extramural Programs
Appropriation Language
For carrying out section 301 and title IV of the PHS Act with respect to health information communications, [$441,997,000]$380,463,000: Provided, That of the amounts available for improvement of information systems, $4,000,000 shall be available until September 30, [2020]2021: Provided further, That in fiscal year [2019]2020, the National Library of Medicine may enter into personal services contracts for the provision of services in facilities owned, operated, or constructed under the jurisdiction of the National Institutes of Health (referred to in this title as “NIH”).
Amounts Available for Obligation 1
(Dollars in Thousands)
| Source of Funding | FY 2018 Final | FY 2019 Enacted | FY 2020 President's Budget |
|---|---|---|---|
| Appropriation | $428,553 | $441,997 | $380,463 |
| Mandatory Appropriation: (non-add) | |||
| Type 1 Diabetes | (0) | (0) | (0) |
| Other Mandatory financing | (0) | (0) | (0) |
| Rescission | 0 | 0 | 0 |
| Sequestration | 0 | 0 | 0 |
| Secretary's Transfer | -1,007 | 0 | 0 |
| Subtotal, adjusted appropriation | $427,546 | $441,997 | $380,463 |
| OAR HIV/AIDS Transfers | 0 | 0 | 0 |
| Subtotal, adjusted budget authority | $427,546 | $441,997 | $380,463 |
| Unobligated balance, start of year | 2,200 | 2,500 | 0 |
| Unobligated balance, end of year | -2,500 | 0 | 0 |
| Subtotal, adjusted budget authority | $427,246 | $444,497 | $380,463 |
| Unobligated balance lapsing | -257 | 0 | 0 |
| Total obligations | $426,989 | $444,497 | $380,463 |
1 Excludes the following amounts (in thousands) for reimbursable activities carried out by this account:
FY 2018 - $7,668 FY 2019 - $7,968 FY 2020 - $7,968
Budget Mechanism Total 1
(Dollars in Thousands)
| MECHANISM | FY 2018 Final | FY 2019 Enacted | FY 2020 President's Budget | FY 2020 +/- FY 2019 Enacted |
||||
|---|---|---|---|---|---|---|---|---|
| No. | Amount | No. | Amount | No. | Amount | No. | Amount | |
| Research Projects: | ||||||||
| Noncompeting | 54 | $20,476 | 60 | $21,984 | 75 | $22,733 | 15 | $749 |
| Administrative Supplements | (4) | 1,041 | (0) | 0 | (0) | 0 | (0) | 0 |
| Competing: | ||||||||
| Renewal | 4 | 2,299 | 4 | 2,200 | 1 | 457 | -3 | -1,744 |
| New | 29 | 8,501 | 34 | 10,360 | 20 | 6,820 | -14 | -3,540 |
| Supplements | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Subtotal, Competing | 33 | $10,801 | 38 | $12,560 | 21 | $7,277 | -17 | -$5,283 |
| Subtotal, RPGs | 87 | $32,319 | 98 | $34,544 | 96 | $30,010 | -2 | -$4,534 |
| SBIR/STTR | 5 | 1,258 | 6 | 1,355 | 6 | 1,125 | 0 | -230 |
| Research Project Grants | 92 | $33,577 | 104 | $35,899 | 102 | $31,135 | -2 | -$4,764 |
| Research Centers: | ||||||||
| Specialized/Comprehensive | 0 | $32 | 0 | $343 | 0 | $242 | 0 | -$101 |
| Clinical Research | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Biotechnology | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Comparative Medicine | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Research Centers in Minority Institutions | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Research Centers | 0 | $32 | 0 | $343 | 0 | $242 | 0 | -$101 |
| Other Research: | ||||||||
| Research Careers | 14 | $2,365 | 13 | $2,182 | 9 | $1,273 | -4 | -$909 |
| Cancer Education | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Cooperative Clinical Research | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Biomedical Research Support | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Minority Biomedical Research Support | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other | 38 | 26,118 | 40 | 25,950 | 34 | 22,611 | -6 | -3,340 |
| Other Research | 52 | $28,483 | 53 | $28,133 | 43 | $23,884 | -10 | -$4,248 |
| Total Research Grants | 144 | $62,092 | 157 | $64,375 | 145 | $55,262 | -12 | -$9,113 |
| Ruth L Kirchstein Training Awards: | FTTPs | FTTPs | FTTPs | FTTPs | ||||
| Individual Awards | 5 | $218 | 5 | $236 | 5 | $239 | 0 | $3 |
| Institutional Awards | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total Research Training | 5 | $218 | 5 | $236 | 5 | $239 | 0 | $3 |
| Research & Develop. Contracts | 0 | $0 | 0 | $0 | 0 | $0 | 0 | $0 |
| (SBIR/STTR) (non-add) | (0) | (3) | (0) | (3) | (0) | (0) | (0) | (-3) |
| Intramural Programs | 615 | 349,726 | 638 | 357,601 | 638 | 307,155 | 0 | -50,445 |
| Res. Management & Support | 85 | 15,510 | 103 | 19,786 | 103 | 17,807 | 0 | -1,979 |
| Res. Management & Support (SBIR Admin) (non-add) | (0) | (0) | (0) | (0) | (0) | (0) | (0) | (0) |
| Construction | 0 | 0 | 0 | 0 | ||||
| Buildings and Facilities | 0 | 0 | 0 | 0 | ||||
| Total, NLM | 700 | $427,546 | 741 | $441,997 | 741 | $380,463 | 0 | -$61,534 |
1 All items in italics and brackets are non-add entries.
Major Changes in the Fiscal Year 2020 President’s Budget Request
Major changes in the FY 2020 President’s Budget request for the National Library of Medicine (NLM), by budget mechanism and/or activity detail are briefly described below. Note that there may be overlap between budget mechanism and activity detail; thus these highlights will not sum to the total change for NLM’s FY 2020 President’s Budget request, which is $380.46 million, a decrease of $61.53 million from the FY 2019 Enacted level. The FY 2020 President's Budget reflects the Administration's fiscal policy goals for the Federal Government. Within that framework and informed by the NLM Strategic Plan, 2017-2027, the NIH Strategic Plan for Data Science, and the 2019 Blue Ribbon Panel review of the NLM Intramural Research Program, the National Library of Medicine will pursue its highest research priorities through strategic investments and careful stewardship of appropriated funds.
Extramural Programs (-$9.110 million; total $55.501 million): With this level of funding, NLM will support the same number of university-based biomedical informatics and data science training programs as under the FY 2019 Enacted level with a reduced funding level. NLM will award up to 20 new research project grants in biomedical informatics and data science and will continue its support of the outreach and stakeholder engagement efforts of the National Network of Libraries of Medicine at a reduced level of funding.
Intramural Programs (-$50.445 million; total $307.155 million): NLM’s intramural programs encompass both intramural research and information service programs that support the acquisition, storage and distribution of biomedical data, the delivery of reliable, high-quality information services, and the development of advanced information systems, standards, and research tools. NLM will maintain funding for intramural research and prioritize research and services that support NIH-wide interests in data science. It will seek ways to streamline and improve the efficiency of its information services while maintaining support for mission critical systems that are heavily used by researchers, clinicians, and the general public. NLM will continue its role as a central coordinating body for the Department of Health and Human Services as relates to standard clinical vocabularies while continuing to seek efficiencies in these programs.
Summary of Changes
(Dollars in Thousands)
| Budget Category | Amount |
|---|---|
| FY 2019 Enacted | $441,997 |
| FY 2020 President's Budget | $380,463 |
| Net change | -$61,534 |
| CHANGES | FY 2020 President's Budget | Change from FY 2019 Enacted | ||
|---|---|---|---|---|
| FTEs | Budget Authority | FTEs | Budget Authority | |
| A. Built-in: | ||||
| 1. Intramural Programs: | ||||
| a. Annualization of January 2019 pay increase & benefits | $104,251 | $322 | ||
| b. January FY 2020 pay increase & benefits | 104,251 | 751 | ||
| c. Paid days adjustment | 104,251 | 0 | ||
| d. Differences attributable to change in FTE | 104,251 | 0 | ||
| e. Payment for centrally furnished services | 3,411 | 0 | ||
| f. Cost of laboratory supplies, materials, other expenses, and non-recurring costs | 199,493 | 0 | ||
| Subtotal | $1,072 | |||
| 2. Research Management and Support: | ||||
| a. Annualization of January 2019 pay increase & benefits | $12,722 | $39 | ||
| b. January FY 2020 pay increase & benefits | 12,722 | 90 | ||
| c. Paid days adjustment | 12,722 | 0 | ||
| d. Differences attributable to change in FTE | 12,722 | 0 | ||
| e. Payment for centrally furnished services | 720 | 0 | ||
| f. Cost of laboratory supplies, materials, other expenses, and non-recurring costs | 4,365 | 0 | ||
| Subtotal | $129 | |||
| Subtotal, Built-in | $1,201 | |||
| CHANGES | FY 2020 President's Budget | Change from FY 2019 Enacted | ||
|---|---|---|---|---|
| No. | Amount | No. | Amount | |
| B. Program: | ||||
| 1. Research Project Grants: | ||||
| a. Noncompeting | 75 | $22,733 | 15 | $749 |
| b. Competing | 21 | 7,277 | -17 | -5,283 |
| c. SBIR/STTR | 6 | 1,125 | 0 | -230 |
| Subtotal, RPGs | 102 | $31,135 | -2 | -$4,764 |
| 2. Research Centers | 0 | $242 | 0 | -$101 |
| 3. Other Research | 43 | 23,884 | -10 | -4,248 |
| 4. Research Training | 5 | 239 | 0 | 3 |
| 5. Research and development contracts | 0 | 0 | 0 | 0 |
| Subtotal, Extramural | $55,501 | -$9,110 | ||
| 6. Intramural Programs | 638 | $307,155 | 0 | -51,518 |
| 7. Research Management and Support | 103 | 17,807 | 0 | -2,108 |
| 8. Construction | 0 | 0 | ||
| 9. Buildings and Facilities | 0 | 0 | ||
| Subtotal, Program | 741 | $380,463 | 0 | -62,736 |
| Total changes | -61,534 | |||
Fiscal Year 2020 Budget Graphs
History of Budget Authority and FTEs:
Bar Graph for Funding Levels by Fiscal Year for FY2016 through FY2020
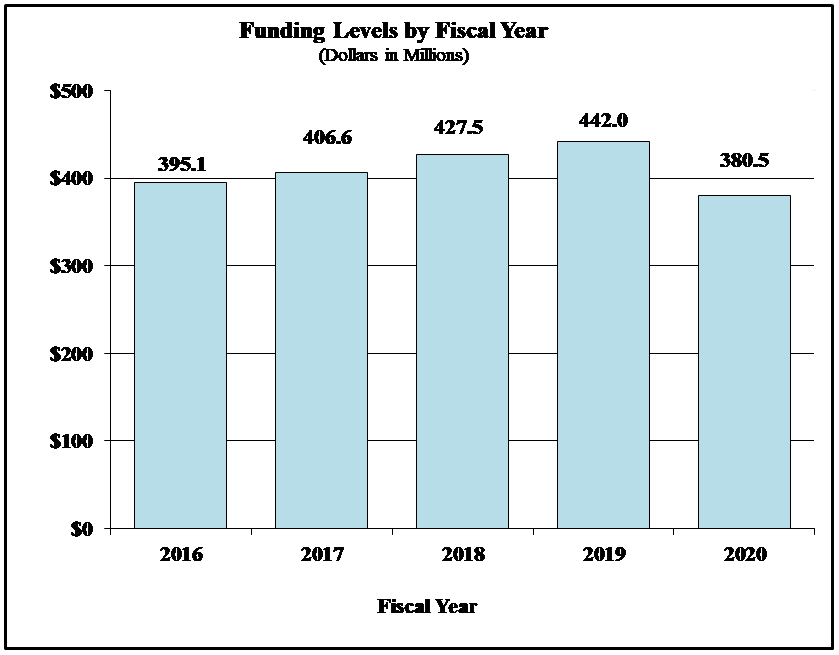
Data for Funding Levels by Fiscal Year for FY2016 through FY2020
| Fiscal Year | Funding (Dollars in Millions) |
|---|---|
| 2016 | 395.1 |
| 2017 | 406.6 |
| 2018 | 427.5 |
| 2019 | 442.0 |
| 2020 | 380.5 |
Bar Graph of FTEs by Fiscal Year for FY2016 through FY2020
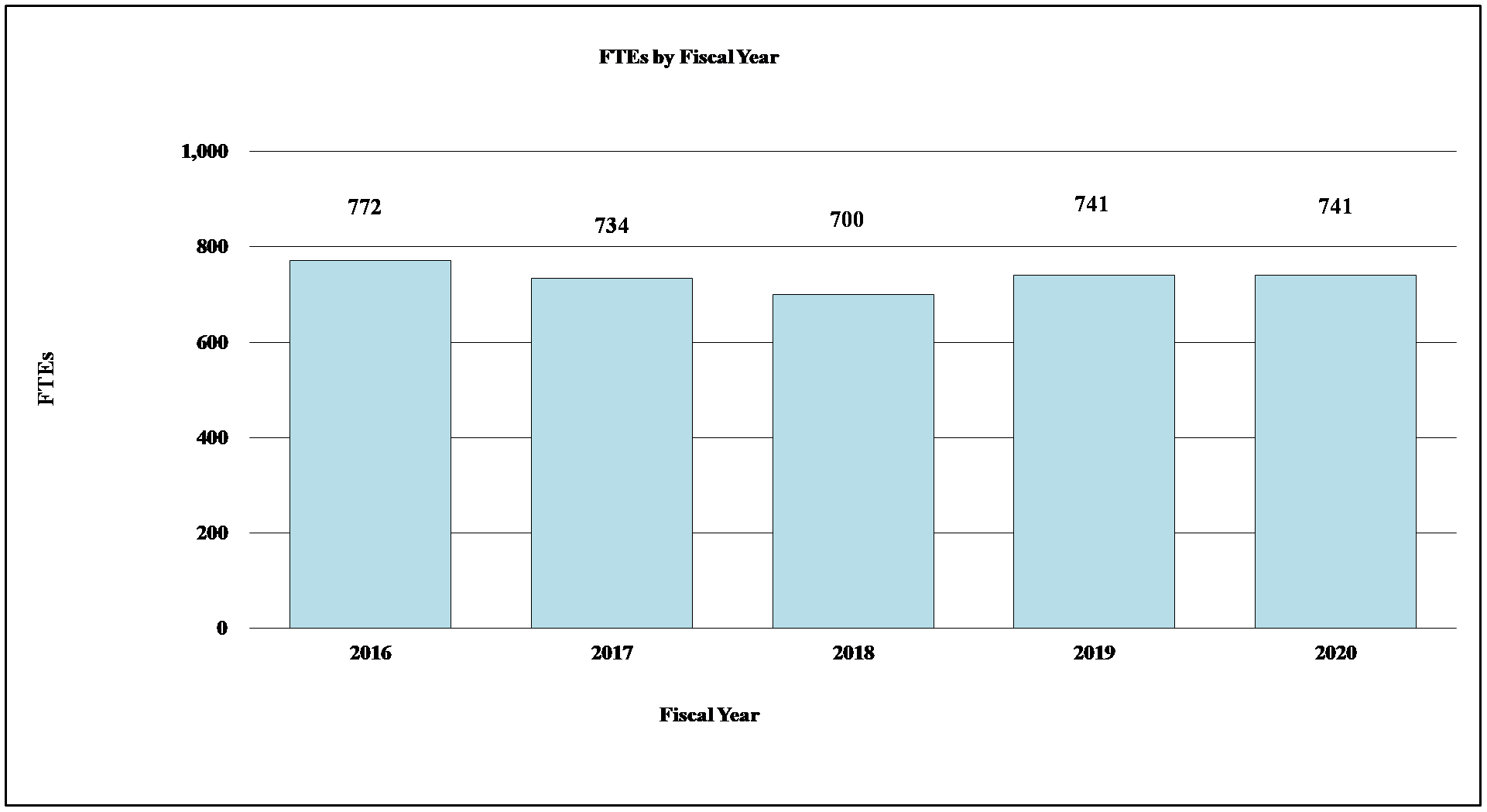
Data for FTEs by Fiscal Year for FY2016 through FY2020
| Fiscal Year | FTEs |
|---|---|
| 2016 | 772 |
| 2017 | 734 |
| 2018 | 700 |
| 2019 | 741 |
| 2020 | 741 |
Distribution by Mechanism:
Pie Chart of FY2020 Budget Mechanisms
(Budget in Thousands)
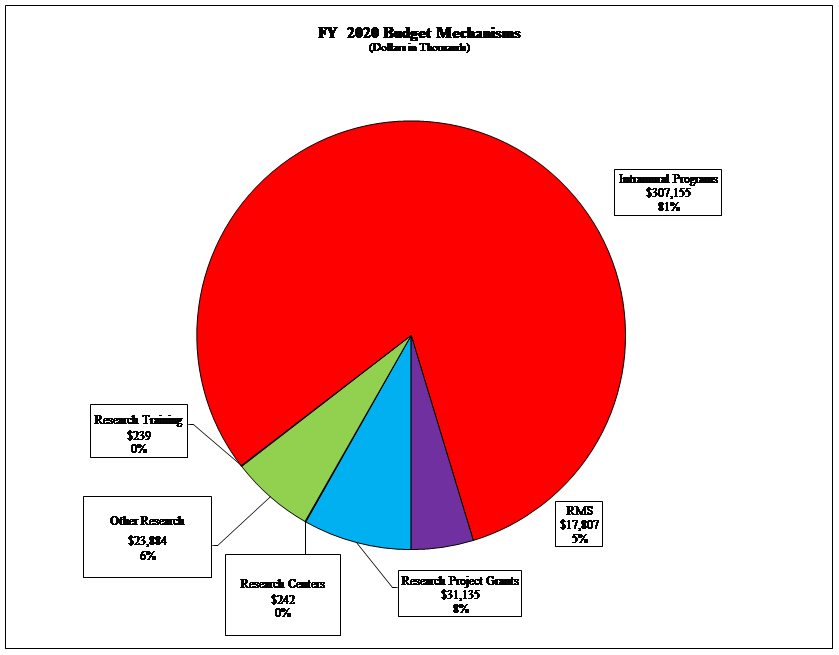
Data for FY2020 Budget Mechanisms
(Budget in Thousands)
| Mechanism | Dollars | Percentage |
|---|---|---|
| Intramural Programs | $307,155 | 81% |
| Research Training | $239 | 0% |
| Other Research | $23,884 | 6% |
| Research Centers | $242 | 0% |
| Research Project Grants | $31,135 | 8% |
| RMS | $17,807 | 5% |
Bar Graph of FY2020 Estimated Percent Change from FY 2019 Mechanism
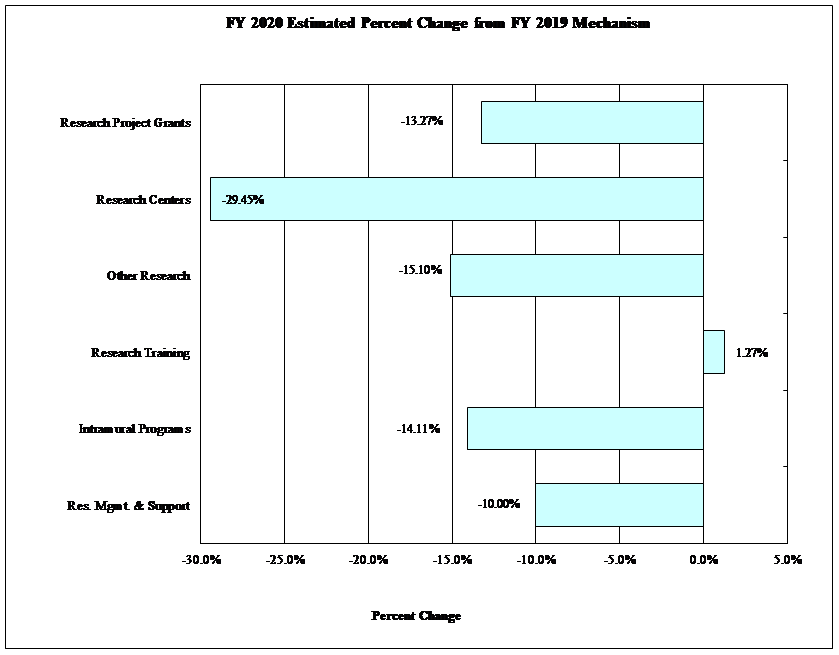
Data for Bar Graph of FY2020 Estimated Percent Change from FY 2019 Mechanism
| Mechanism | Percentage Change |
|---|---|
| Intramural Programs | -14.11% |
| Research Training | 1.27% |
| Other Research | -15.10% |
| Research Centers | -29.45% |
| Research Project Grants | -13.27% |
| Res. Mgmt. & Support | -10.00% |
Budget Authority By Activity 1
(Dollars in Thousands)
| FY 2018 Final | FY 2019 Enacted | FY 2020 President's Budget | FY 2020 +/- FY2019 Enacted |
|||||
|---|---|---|---|---|---|---|---|---|
| Extramural Research | FTE | Amount | FTE | Amount | FTE | Amount | FTE | Amount |
| Detail | ||||||||
| Health Information for Health Professionals and the Public (NN/LM) | $11,911 | $11,911 | $9,239 | -$2,672 | ||||
| Informatics Resources for Biomedicine and Health | 16,824 | 16,457 | 15,126 | -1,332 | ||||
| Biomedical Informatics Research | 33,574 | 36,242 | 31,135 | -5,107 | ||||
| Subtotal, Extramural | $62,310 | $64,611 | $55,501 | -$9,110 | ||||
| Intramural Programs | 615 | $349,726 | 638 | $357,601 | 638 | $307,155 | 0 | -$50,445 |
| Research Management & Support | 85 | $15,510 | 103 | $19,786 | 103 | $17,807 | 0 | -$1,979 |
| TOTAL | 700 | $427,546 | 741 | $441,997 | 741 | $380,463 | 0 | -$61,534 |
1Includes FTEs whose payroll obligations are supported by the NIH Common Fund.
Authorizing Legislation
| PHS Act/ Other Citation |
U.S. Code Citation |
2019 Amount Authorized |
FY 2019 Enacted | 2020 Amount Authorized |
FY 2020 President's Budget | |
|---|---|---|---|---|---|---|
| Research and Investigation | Section 301 | 42§241 | Indefinite | $441,997,000 | Indefinite | $380,463,000 |
| National Library of Medicine | Section 401(a) | 42§281 | Indefinite | |||
| Indefinite | ||||||
| Total, Budget Authority | $441,997,000 | $380,463,000 | ||||
Appropriations History
| Fiscal Year | Budget Estimate to Congress | House Allowance | Senate Allowance | Appropriation |
|---|---|---|---|---|
| 2011 | $364,802,000 | $364,254,000 | $339,716,000 | |
| Rescission | $2,982,909 | |||
| 2012 | $387,153,000 | $387,153,000 | $358,979,000 | $338,278,000 |
| Rescission | $639,345 | |||
| 2013 | $372,651,000 | $381,981,000 | $337,638,655 | |
| Rescission | $675,277 | |||
| Sequestration | ($16,947,139) | |||
| 2014 | $382,252,000 | $387,912,000 | $327,723,000 | |
| Rescission | $0 | |||
| 2015 | $372,851,000 | $336,939,000 | ||
| Rescission | $0 | |||
| 2016 | $394,090,000 | $341,119,000 | $402,251,000 | $394,664,000 |
| Rescission | $0 | |||
| 2017¹ | $395,684,000 | $407,086,000 | $412,097,000 | $407,510,000 |
| Rescission | $0 | |||
| 2018 | $373,258,000 | $413,848,000 | $420,898,000 | $428,553,000 |
| Rescission | $0 | |||
| 2019 | $395,493,000 | $433,671,000 | $442,230,000 | $441,997,000 |
| Rescission | $0 | |||
| 2020 | $380,463,000 |
Justification of Budget Request
Authorizing Legislation: Section 301 and title IV of the Public Health Service Act, as amended.
Budget Authority (BA):
| FY 2018 Actual |
FY 2019 Annualized CR |
FY 2020 President’s Budget |
FY 2020 +/- FY 2019 |
|
|---|---|---|---|---|
| BA | $427,546,000 | $441,997,000 | $380,463,000 | -$61,534,000 |
| FTE | 700 | 741 | 741 | 0 |
Program funds are allocated as follows: Competitive Grants/Cooperative Agreements; Contracts; Direct Federal/Intramural and Other.
Director's Overview
Through its cutting-edge research, information systems, collections, and training programs, NLM plays an essential role in catalyzing basic biomedical science and data-driven discovery. NLM acquires, organizes, curates, and delivers up-to-date biomedical information across the United States and around the globe. Millions of scientists, health professionals, and members of the public use NLM’s electronic information sources millions of times each day. NLM makes research results available for translation into new treatments, products, and practices; provides decision support for health professionals and patients; and aids disaster and emergency preparedness and response.
NLM is a leader in data science and open science, including the acquisition and analysis of data for discovery and the training of biomedical data scientists. As biomedical research becomes increasingly digital, NLM is expanding its data science and biomedical informatics research programs to improve access to digital knowledge assets, including research and bibliographic data, software used to generate or analyze research data, and models and workflows used in research. Making such materials findable, accessible, interoperable, and re-usable (FAIR), as well as attributable and sustainable, accelerates science, broadens opportunities for collaboration, enhances accountability (e.g., enabling and assessing reproducibility,) and increases the return on investments in research for research participants, science, and society.
Priorities for FY 2020
The NLM Strategic Plan 2017-2027 positions NLM as a platform for biomedical discovery and data-powered health through the provision of tools for data-driven research, reaching more people in more ways through enhanced dissemination and engagement pathways, and building a workforce for data-driven research and health. In implementing its Strategic Plan, NLM strives to systematically characterize and curate data reflecting the full spectrum of health phenomena from cells to individuals to society, and to devise new methods for extracting knowledge from such data. While the majority of NLM’s budget is allocated to products and services that make data and literature widely available, NLM aims to expand its intramural and extramural research programs, with a particular emphasis on advancing the applications of machine learning and new approaches to computational biology.
NLM is also addressing existing and emerging challenges in biomedical research and public health by expanding its core biomedical literature and genomic collections to include a broad array of health and biological data types and making these data FAIR. Additionally, NLM will facilitate implementation of the NIH Strategic Plan for Data Science through advanced research in biomedical informatics and data science, technical contributions to the development of sustainable data infrastructure, training resources for NIH staff, and contributions to policy efforts for improving the preservation of and access to data from NIH-funded research. NLM will continue to foster data stewardship through its commitment to sustainable technical infrastructure, including the appropriate use of commercial cloud resources. NLM will expand its training programs to include an increased focus on data science, expand its support of a diverse workforce, and maintain its commitment to outreach excellence.
Basic Computational Biology and Biomedical Science Research
NLM maintains a robust research program in computational health information science. Scientists around the world analyze data and information using software, algorithms, and methods arising from basic research in biomedical informatics, data science, and computational biology conducted or funded by NLM. NLM research emphasizes new methods and approaches to foster data-driven discovery in the biomedical and clinical health sciences, as well as domain-independent, reusable approaches to discovery, curation, analysis, organization and management of health-related digital objects.
NLM research in machine learning, deep learning, and natural language processing has improved and accelerated the diagnosis and treatment of diseases ranging from cervical cancer to tuberculosis to macular degeneration. It has also enabled identification of previously unknown adverse drug reactions and assisted in identifying patients for potential inclusion in clinical research. NLM-funded informatics and data science research has led to exciting innovations in mining data from electronic health records (EHRs) and social media to improve prediction and assessment of influenza and foodborne illnesses in real-time.
NLM’s research has led to insights in the role of genetic factors and mutational patterns in disease, gene regulation, molecular binding, and protein structure and function, as well as new methods and approaches that have enabled characterizing the clustered regularly interspaced short palindromic repeats, CRISPR-associated genes (CRISPR-Cas) system, discoveries in cancer genomics, and large scale characterization of genomic control elements.
NLM research has also enabled production resources for automatic recognition and annotation of virus and structural RNA sequences, which has led to the ability to assemble huge numbers of complete bacterial genomes and automatically annotate and classify them, summarizing and condensing raw data into actionable reports used for food pathogen surveillance by the Food and Drug Administration (FDA) and Centers for Disease Control and Prevention (CDC), and populating NLM’s public sequence research databases.
Exploring the Next Frontier in Data Science
NLM aims to strengthen its position as a center of excellence for health data analytics and discovery, as well as to spearhead application of advanced data science tools to biological, clinical, and health data. To support knowledge discovery, NLM develops and applies statistical and machine learning methods to predict outcomes, and generates evidence for clinical decision support and precision medicine. NLM engages in multi-stakeholder, interdisciplinary collaboratives to bring out the value of data through large-scale analytics for drawing consistent, actionable and correct inferences from such data and for extracting relevant characteristics from clinical images and text. In collaboration with other NIH Institutes and Centers (ICs), NLM develops and uses methods and test sets for processing narrative text; and develops and studies deep learning methods and other techniques for analyzing and classifying different types of biomedical images.
NLM will continue to conduct activities to facilitate implementation of NIH’s Strategic Plan for Data Science, which includes supporting new models for data stewardship. NLM is also strengthening its contributions to high-priority NIH and HHS initiatives, such as the All of Us Research Program, the BRAIN Initiative, and efforts dedicated to address the opioid crisis, HIV/AIDS, and Alzheimer’s disease.
A Workforce for Data-Driven Research and Health
NLM is committed to building a workforce for data-driven research and health. NLM continues to promote and fund PhD-level research training in biomedical informatics and data science. In addition, the NLM will partner across the NIH to ensure inclusion of data science and open science core skills in all NIH training programs, and will expand training for librarians, information science professionals, and other research facilitators. Such efforts will help to ensure data science and open science proficiency, expand research methods which support rigor and reproducibility, promote and increase workforce diversity, and engage the next generation of researchers. To address the lack of diversity that exists in these fields, NLM has partnerships with high schools, information schools, and minority-serving institutions. Further, NLM is participating in NIH-wide efforts to foster a culture that not only advances science, but also ensures the development and retention of a diverse, safe, and respectful workforce for the future.
Transformational Tools and Technologies
NLM builds information products and services that support biomedical research, clinical care, and consumer health. Its richly-linked databases promote scientific breakthroughs, playing an essential role in all phases of research and innovation. Every day, NLM serves more than 5 million users, receiving up to 15 terabytes of new data, adding value by enhancing data quality and consistency, and integrating new data with other information in NLM databases. NLM responds to millions of inquiries per day from individuals and computer systems and provides more than 115 terabytes of information, including more than 5.1 million published articles in PubMed Central (PMC), NLM’s free full-text archive of biomedical and life sciences journal literature. NLM creates resources, tools, technologies, and services for literature, data, and standards that drive innovation and discovery and improve health, thus enabling the application of knowledge at the point of care. NLM brings the power of data science to citizens and makes its resources available to users where and when they are most needed through tools such as mobile apps and application programming interfaces (APIs) which enable machine-to-machine use of data and information. NLM-funded research also supports the development of tools which enable consumers to better organize and maintain personal libraries of relevant health information.
NLM researchers devise ways to make the delivery of NLM services more efficient and capable of handling ever-growing amounts of data. NLM’s curation activities play an important role in ensuring data quality and the public’s trust in NLM’s tools and resources; they are being increasingly automated. Genomic pipeline processing for NLM’s Pathogens Project for foodborne pathogens is accelerated to allow for more rapid first response to process submissions within 24 hours. NLM continues to make improvements to the PubMed system – its widely-used search engine for citations to biomedical literature – to automatically characterize what users would consider the critical elements in biomedical journal articles, extract drug, disease, and gene information. These improvements also allow PubMed to deliver the most relevant information to users, applying machine learning to create relevance-based rankings of search result and more prominent links to associated data and data citations.
With new developments and enhancements to its systems, NLM will strive to provide access to its resources on any platform, including mobile devices. NLM is committed to citizen science and will continue to offer events, such as hackathons (hands-on, team-based training projects), that welcome community members to work with NLM staff to improve its products and services.
Looking to the future, NLM will build on its strong scientific tradition, continuing to employ cutting edge computational and artificial intelligence (AI) approaches to conduct validation and ensure statistical rigor and taking advantage of cloud-based, federated computing architectures. In doing so, NLM will develop automated methods of extracting information from more types of data, both to make new biomedical discoveries and disseminate new analytical approaches. It will also leverage knowledge gained from its research to improve and automate data submission processes, make analytical tools and pipelines available to users, and improve linkages among datasets or to the published record.
Overall Budget Policy
The FY 2020 President’s Budget request is $380.463 million, a decrease of $61.534 million from the FY 2019 Enacted level for NLM. Funds are included to advance intramural research in computational health information sciences and methods, such as artificial intelligence and machine learning, and maintain NLM’s most heavily used information services that provide access to published biomedical literature and consumer health information. At this level, support for critical NLM databases and services will be prioritized, ensuring mission critical support for NIH data science goals and data sharing policies. NLM will support key intramural data science activities, including processing and organizing genomic data resulting from NIH-wide investments in whole genome sequencing; evaluation of microbial and viral pathogens to support outbreak detection and response by CDC, FDA, and USDA; enhancing registration and results reporting in ClinicalTrials.gov; and updating and disseminating clinical terminology standards required for interoperability of U.S. health data. NLM will also award up to 20 new research project grants through its extramural programs. NLM will continue to leverage the National Network of Libraries of Medicine and expand its role to advance community engagement in the All of Us initiative. NLM will prioritize and consolidate outreach programs that promote access and training in the effective use of NLM resources for scientists, clinicians, patients, and the general public.
Program Descriptions and Accomplishments
Intramural Program
NLM’s intramural program supports research and training in computational health information science, drawing upon the disciplines of biomedical informatics, data science, and computational biology. It also supports the development and operation of high-value biomedical information services and systems, including for health data standards. As part of these efforts, NLM invests in outreach and training to help researchers, clinicians, educators, and the public learn about and make effective use of its wide range of health information services.
Intramural Research and Training
NLM’s intramural research program develops and applies computational approaches to a broad range of problems in biomedicine, molecular biology, and health. With components housed in two Congressionally-chartered centers, the Lister Hill National Center for Biomedical Communications and the National Center for Biotechnology Information, the program brings together scientific personnel with expertise in computer science, library science, engineering, biochemistry, molecular biology, mathematics, statistics, structural biology, medicine, and related disciplines, to develop and apply novel computational, informatics, and data science approaches to improve biomedicine. It advances computational biomedical discovery, develops and applies novel algorithms to image interpretation and clinical records analysis, contributes to informatics and data science needs of the broader biomedical research community, and improves NLM’s advanced information services.
Health Data Powered Discovery.
NLM data science, informatics research, and innovative clinical analytic methods enable researchers to query large sets of patient data across institutional boundaries, conduct predictive clinical analytics, and answer important biomedical research questions through observational and pragmatic clinical trial approaches. For example, in FY 2018, NLM provided technical expertise and assistance for trans-NIH authentication and authorization to research data hosted on multiple cloud providers in support of NIH-wide initiatives in data science. NLM also provides technical leadership into activities that support the NIH New Models of Data Stewardship program, including the NIH Strategic Plan for Data Science and the Science and Technology Research Infrastructure for Discovery, Experimentation, and Sustainability (STRIDES) Initiative.
Machine Learning.
NLM’s intramural research program has a strong focus on development and application of machine learning to improve the diagnosis and treatment of a wide range of diseases, often in collaboration with other NIH ICs. For example, NLM researchers worked with the National Cancer Institute (NCI) to develop deep learning models to aid in interpreting cervical images to detect the presence of human papillomavirus infection (a cause of cervical cancer), improve classification of the disease, and assist in early treatment in low resource areas. In addition, NLM researchers developed a highly reliable deep learning algorithm that screens for malaria with 99 percent accuracy by detecting the presence of the malaria-causing parasite (plasmodium falciparum) in red blood cell images. The algorithm, implemented in a smartphone, underwent successful field tests in low-resource areas and has broad applicability to abnormal blood cell recognition. NLM also conducted deep learning research on detecting and classifying pneumonia in pediatric chest x-rays in children ages one to five years. The result was a highly accurate pneumonia detection algorithm that could aid in diagnosis and treatment by distinguishing between bacterial and viral pneumonia with 90 percent sensitivity and 96 percent specificity.
Text Mining.
NLM researchers develop and test advanced text mining techniques that can be used, for example, to improve the retrieval of biomedical information from NLM databases and analyze unstructured clinical text in EHR data. Results of work in FY 2018, including collaborating with the National Institute on Drug Abuse (NIDA) to study the adverse effects of opioid use in the elderly, demonstrated the potential of natural language processing techniques to enable broader use of EHRs as a source of data for clinical research and natural history studies, to evaluate the feasibility of a clinical trial design, and to assess adverse events and risks associated with drug exposure in post-market observational studies.
Computational Biology.
NLM research in computational biology applies advanced computational approaches to a broad range of fundamental problems in evolution, molecular biology, genomes, biomedical science, and bioinformatics. For example, in FY 2018, NLM researchers:
- Conducted research on methods, measures, and statistics for the comparison and analysis of DNA and protein sequences, automated construction of hierarchical multiple alignments and Hidden Markov Models for elucidating protein subfamily functions and their sequence determinants. NLM also developed and made freely available the LogOddsLogo program for other researchers to use to conduct such analysis.
- Completed an integrated genome-wide analysis of expression for quantitative trait loci that aids the interpretation of genomic association studies. This complex multi-year, multi-institute collaboration characterized genetic variants in the Framingham Heart Study population that are associated with the differential expression of protein coding genes, enabling new uses of this well-known Federal investment for further research purposes.
- Developed a new method to extract from large sets of patient data (most recently cancer cell mutations) gene modules characterized by specific mutational patterns. The method investigates various aspects of cancer cell mutations, enabling discovery of new relationships between mutated gene modules, cancer subtypes, and mutational signatures, and is contributing to a better understanding of the mutational landscape of cancer. The detection of mutational patterns and identification of gene subtypes allows for a better understanding of the trends of cancer mutations in individuals.
- Used sensitive sequence and structure analysis together with comparative genomics and other computational data visualization techniques to study the evolution and biochemistry of proteins and infer organismal biology. This work led to the discovery of enzymes responsible for epigenetic oxidative DNA modifications, a unified mechanism for nucleotide-based regulation of animal immune response and prokaryotic CRISPR systems, and novel toxin systems deployed in inter-organismal biological conflicts.
- Developed computational methods for identification of CRISPR-Cas loci and comprehensive genome analysis leading to the classification of CRISPR-Cas microbial adaptive immune systems. This work identified a new class of CRISPR-Cas proteins, and has served to advance the field of gene editing.
Research training and citizen science.
NLM’s intramural research program provides opportunities for post-doctoral training and career development, as well as opportunities for high-school, college, graduate students, medical professionals, and visiting scientists on sabbatical. In FY 2018, NLM supported 63 fellows in short-term and multi-year training. NLM aims to expand intramural training in biomedical informatics and data science, and improve outreach to recruit diverse and highly-qualified trainees. NLM is also committed to encouraging citizen science as a way to provide opportunities for members of the community to work with NLM to improve and apply NLM products and services in novel ways. In part, NLM accomplishes this through community-based hackathons, which are hands-on, team-based training projects that engage community members to work with NLM staff to improve its products and services. In FY 2018, NLM conducted 14 hackathons on topics such as novel virus discovery and reproducible bioinformatics platforms.
Delivery of High Quality, Trusted Biomedical and Health Information Services
NLM continues to expand the quantity and range of high-quality information readily available to scientists, health professionals, and the public. Its heavily used information services span the gamut from biomedical literature to data to health data standards. NLM continues to seek efficiencies and improvements in its information services through automation, integration of platforms, and improved user interfaces. NLM created PubMed Labs, a test site for experimentation with new features and tools that could be incorporated into PubMed. NLM continues to improve procedures for selecting and curating information sources to provide the public with reliable, trusted health data and information. For example, in FY 2018, NLM:
- Added more than 1.2 million new citations in FY 2018 to PubMed, NLM’s most heavily used database, which contained records for more than 29.1 million life science and biomedical journal articles at the end of CY 2018. Of those citations added in FY 2018, 904,934 citations were indexed for MEDLINE, the largest subset of PubMed citations, using NLM’s controlled vocabulary, Medical Subject Headings (MeSH). Indexing adds value to the citation and abstract by adding subject headings, classifying certain types of research (e.g., clinical trial, systematic review), and improving links to related information (e.g., data sets). NLM also launched a 5-year initiative to explore novel approaches to automated indexing.
- Added approximately half a million full-text articles to PMC, which now includes more than 5.1 million research articles, including those produced by researchers funded by NIH, 10 other Federal agencies, and private research funders. On a typical day, more than 2.5 million users download articles from PMC. In FY 2018, NLM enhanced procedures to accept small datasets related to articles in PMC to facilitate their discovery and re-use.
- Grew ClinicalTrials.gov, the world’s largest clinical trials registry, which now contains approximately 287,000 registered studies and summary results for more than 33,000 studies. In addition, NLM enhanced the availability and display of information in ClinicalTrials.gov to assist users in finding and interpreting search results.
- Expanded the number of topics covered in MedlinePlus, NLM’s source of consumer-oriented health information, including the addition of laboratory test information. MedlinePlus Connect provides patients with a way to link to trusted, consumer-oriented information about diagnoses and drugs directly from their EHR. In 2018, NLM fulfilled more than 156 million queries and delivered information to patients and health care providers at the point of need.
- Integrated the ToxNet suite of toxicology databases with NLM’s other chemical information system, PubChem, on a common technical platform with other NLM electronic resources to improve discoverability and interoperability through common search terminologies and APIs.
- Enhanced NLM’s Wireless Information System for Emergency Responders (WISER), a mobile application that brings important toxicology resources and health information to first responders before, during, and after emergencies, without the need for wireless Internet connections.
- Improved support of device safety information through expansion of NLM’s Access Global Unique Device Identification (Access GUDID) database, which contains key device identification information submitted to the Food and Drug Administration (FDA), and grew 25 percent in FY 2018, with unique identifiers and registration information for more than 1.8 million medical devices.
- Expanded the database of Genotypes and Phenotypes (dbGaP) which connects individual-level genomic data with individual-level clinical information, by 23 percent to more than 1,100 studies involving more than 2 million people. dbGaP provides access to studies that investigate the interaction between genotypes and phenotypes, for both individual-level genomic data and genomic summary results. Following changes to data management procedures under the NIH Genomic Data Sharing Policy, dbGaP now allows unrestricted access to summary-level genomic results from most NIH-supported genomic studies.
- Expanded the Database of Single Nucleotide Polymorphisms (dbSNP), NLM’s database of all known genetic variants of a given size and location, to 672 million records. Also expanded ClinVar, a public archive of reports of the relationships among human genetic variations and phenotypes, to 468,000 records of unique variations, a 34 percent growth in submissions in FY2018.
- Grew NLM’s RefSeq database of integrated, non-redundant, well-annotated reference genomic sequences, which are essential to identifying and documenting genetic variations that affect human health, to over 173 million records in FY 2018, a 23 percent increase.
- Continued to enhance its computing technology infrastructure by taking advantage of advanced computing techniques, such as cloud storage, which offer greater agility, better resource utilization, and potentially lower cost. NLM is exploring advanced cloud implementations of several of its resources, including Access GUDID and PubMed Labs.
- Added more than 3,000 digitized printed texts and born-digital documents to its Digital Collections, a free online archive of biomedical texts, images, and audiovisuals.
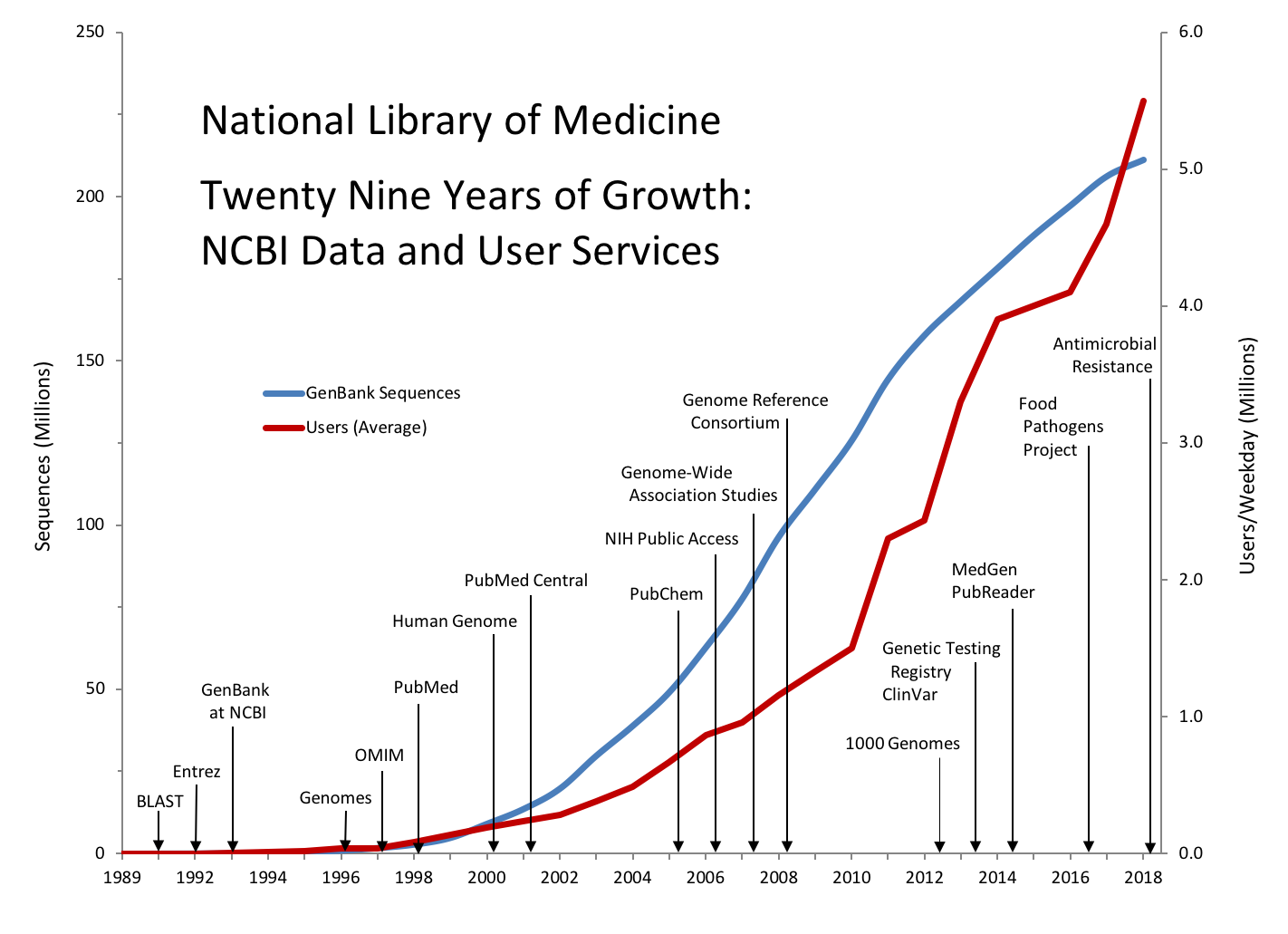
Data for Twenty Nine Years of Growth: NCBI Data and User Services
| Year | Product Release | GenBank Sequences | Users (Average) |
|---|---|---|---|
| 1989 | 30,010 | 1,200 | |
| 1990 | BLAST | 40,295 | 1,500 |
| 1991 | Entrez | 58,952 | 2,700 |
| 1992 | GenBank at NCBI | 87,846 | 3,200 |
| 1993 | 143,492 | 4,500 | |
| 1994 | 215,273 | 9,000 | |
| 1995 | Genomes | 555,694 | 19,000 |
| 1996 | OMIM | 1,021,211 | 36,000 |
| 1997 | PubMed | 1,765,847 | 39,268 |
| 1998 | 2,837,897 | 84,256 | |
| 1999 | Human Genome | 4,864,570 | 138,139 |
| 2000 | PubMed Central | 9,102,634 | 190,485 |
| 2001 | 13,602,262 | 233,977 | |
| 2002 | 19,808,101 | 283,590 | |
| 2003 | 29,819,397 | 380,104 | |
| 2004 | PubChem | 38,941,263 | 485,029 |
| 2005 | NIH Public Access | 49,152,445 | 666,917 |
| 2006 | Genome-Wide Association Studies | 62,765,195 | 864,586 |
| 2007 | Genome Reference Consortium | 77,632,813 | 958,584 |
| 2008 | 96,400,790 | 1,152,700 | |
| 2009 | 110,946,879 | 1,328,143 | |
| 2010 | 1000 Genomes | 125,764,384 | 1,501,335 |
| 2011 | 144,458,648 | 2,300,000 | |
| 2012 | Genetic Testing Registry and ClinVar | 157,889,737 | 2,430,000 |
| 2013 | MedGen and PubReader | 168,335,396 | 3,300,000 |
| 2014 | Pubmed Commons | 178,322,253 | 3,900,000 |
| 2015 | Food Pathogens Project | 188,372,017 | 4,000,000 |
| 2016 | 197,390,691 | 4,100,000 | |
| 2017 | 206,293,625 | 4,600,000 | |
| 2018 | Antimicrobial Resistance | 211,281,415 | 5,500,000 |
Program Portrait: NLM Support of Health Information and Data Standards
FY 2019 Level: $ 19.00 million
FY 2020 Level: $ 15.00 million
Change: -$ 4.00 million
As the coordinating body for clinical terminology standards within the Department of Health and Human Services, NLM maintains a diverse portfolio of products and services which supports interoperability of health data. In close collaboration with the Office of the National Coordinator for Health Information Technology and other Federal departments and agencies, NLM develops, funds, and disseminates the clinical terminologies and coding systems designated as U.S. standards to promote interoperability and health information exchange. NLM develops RxNorm for identifying drugs; and supports the development of LOINC (Logical Observation Identifiers Names and Codes) for identifying tests, measurements and observations, and SNOMED CT (Systematized Nomenclature of Medicine -- Clinical Terms) for identifying problems, organisms, and many other special items. NLM’s support enables key standards to be used free-of-charge in the United States, which promotes data exchange and interoperability to advance health care, public health, biomedical research, and product development.
NLM also supports the development, maintenance, and distribution of the tools and resources needed for implementation of health data standards. This includes the Value Set Authority Center for reporting the Centers for Medicare & Medicaid Services (CMS) e-Clinical Quality Measures, Access GUDID for key device identification information submitted to the FDA about medical devices, and the NIH Common Data Elements Repository for providing access to standard variables and measures to be used in NIH-funded clinical research. In FY 2018, NLM successfully migrated to a new set of tools for authoring, mapping, content requests, and production of the U.S. edition of SNOMED CT, increasing efficiency and productivity while reducing maintenance costs. NLM also developed RxNav, a new graphical user interface and APIs to facilitate access to its drug terminologies by researchers, industry, and the public. In FY 2018, use of these drug APIs reached a new high of one billion queries. NLM also added new attributes to better align RxNorm with global drug information standards to improve global interoperability for medicinal products.
Public Awareness and Access to Information
NLM uses multiple channels to reach the public, including direct contact, development of consumer-friendly websites, and human networks that reach out to communities. In FY 2018, NLM and the 7,100 members of the National Network of Libraries of Medicine (NNLM) funded more than 300 outreach projects to enhance awareness of and access to NLM’s health information services. Working with community organizations, academic institutions, American Indian tribes, health professionals, and librarians, NLM and NNLM addressed the opioid crisis, health disparities in diverse communities, environmental health issues, women’s health, HIV/AIDS, and more. In FY 2018, NLM and the NNLM worked together to ensure the general public, emergency medicine, and public health professionals have access to information needed to address a variety of health needs following wildfires, hurricanes, and floods.
In partnership with NIH ICs and other collaborators, NLM produces the quarterly NIH MedlinePlus magazine, and its Spanish counterpart, NIH Salud. The magazine is distributed to 70,000 individual and bulk subscribers, as well as to doctors’ offices, health science libraries, Congress, the media, federally supported community health centers, select hospital emergency and waiting rooms, and other locations nationwide where the public receives health services. In FY 2018, NLM developed a new online version of the NIH MedlinePlus magazine to reach more people in more ways through enhanced dissemination and engagement.
NLM uses exhibitions, the media, and new technologies in its efforts to reach under-served populations and promote interest among young people in science, medicine, and technology. NLM continues to expand its successful traveling exhibitions program, which is a cost-efficient way to extend access to NLM resources around the world, including international locations for access by U.S. Armed Forces. In FY 2018, public-private partnerships enabled NLM traveling exhibitions to appear in 181 institutions in 161 towns and cities in 44 states and two other countries.
Budget Policy
The FY 2020 President’s Budget estimate for NLM’s Intramural Programs is $307.155 million, a decrease of $50.445 million from the FY2019 Enacted Budget of $357.601 million. NLM’s FY 2019 expenditure for intramural research was approximately $27 million and will remain unchanged in FY 2020. NLM’s intramural research program advances knowledge in data science methods, including artificial intelligence and machine learning, as well as the advanced integration and analysis of genomic data, clinical research data, and observational health data. Consistent with recommendations from the Blue Ribbon Panel review of its intramural research program completed in FY 2019, NLM will align its intramural research activities under a single Scientific Director, seek efficiencies in core research services, and hire two intramural investigators. Research priorities will be informed by NLM’s long tradition of conducting research that informs effective delivery of NLM services, including research in health data standards development, use, and evaluation; computational approaches to curation; and machine-learning approaches to text and image processing.
NLM will support key intramural data science activities, including automated indexing and management of the biomedical literature, processing and organizing genomic data resulting from NIH-wide investments in whole genome sequencing, and genomic evaluations of pathogens in support outbreak detection and response by CDC, FDA, and USDA. The Library will prioritize and seek new efficiencies in its service offerings to ensure needed levels of support for biomedical and health information resources that are most heavily used by the scientific research community and the public and that support NIH-wide data science goals and data sharing policies. NLM will support further enhancements to ClinicalTrials.gov to facilitate submission of and access to clinical trial data that must be submitted in accordance with the Food and Drug Administration Amendments Act and NIH policy. It will improve tools for updating and disseminating clinical terminology standards required for interoperability of data contained in EHRs and to support advanced integration and analysis of genomic, clinical research, and observational health data
In FY 2020, NLM will prioritize its outreach programs within its Intramural Program to improve its efforts to promote access and training in the effective use of NLM resources, including data repositories, and to increase engagement with broad sets of stakeholders in the public and private sectors to leverage their talents, capabilities, and information resources. Through its cooperative agreement with the National Network of Libraries of Medicine, NLM will continue to support efforts to advance community engagement in the All of Us initiative.
Program Portrait: National Network of Libraries of Medicine (NNLM)
FY 2019 Level: $ 11.13 million
FY 2020 Level: $ 9.24 million
Change: -$ 1.89 million
The 7,100 member institutions of the NNLM are valued partners in ensuring that health information, including from NLM services, is available to scientists, health professionals, and the public. NNLM is comprised of eight Regional Medical Libraries (RMLs), each anchoring a regional consortium of academic health sciences libraries, hospital libraries, public libraries, and community-based organizations. Five national offices serve as central infrastructure and program service points. The RMLs and Offices are supported through cooperative agreements awarded to health sciences libraries at the University of California, Los Angeles; University of Iowa; University of Maryland; University of Massachusetts; University of North Texas; University of Pittsburgh; University of Utah; and University of Washington.
The NNLM has a partnership with the NIH All of Us Research Program to support community education and engagement activities designed to raise awareness about the program to library audiences. The partnership also provides a learning platform for and about All of Us for the program’s entire community, including participants, researchers, program providers, and partners. In FY 2018, the NNLM formed the All of Us Community Engagement Network (CEN), which is the subset of NNLM members who support, promote, and lead All of Us engagement activities in libraries. The main goals are to increase capacity for public library staff to provide health information services for their patrons and to blanket the nation with All of Us awareness through library activities. As of December 2018, the NNLM partnership with the NIH All of Us Research Program has reached 16 states.
Public library systems in Albuquerque, New Orleans, Durham, Memphis, and Nashville were initial CEN members. They provide services ranging from mobile outreach through book-mobiles and mobile technology units featuring the All of Us Research Program to health and digital literacy training that prepares participants, especially from historically underrepresented populations in biomedical research, to better use information received from All of Us.
To build capacity and raise awareness of All of Us, the NNLM provided in-depth consumer health information training to over 300 public library staff in FY 2018, exceeding the annual target of 200. It also ensured a comprehensive library presence in the May 2018 program launch. For the remainder of FY 2018, the CEN conducted more than 150 events for more than 500 participants, including presentations, workshops, and exhibits introducing the program to library staff and audiences ranging from Tribal College Librarians Institute to the Allied Media Conference.
The NNLM’s All of Us Training and Education Center (TEC) operates the program’s learning platform. In FY 2018, the platform was launched with an initial training offering required for new program providers. Future developments include training programs for researcher ethics and educational materials supporting the return of genomic results to participants. The TEC also employs education as an engagement strategy for participants. In FY 2019, TEC-designed learning activities and educational experiences will be deployed for use in library activities across the country as part of a monthly series tied to National Health Observances.
Extramural Programs
NLM’s extramural programs support the development of knowledge, infrastructure, and human resources needed to advance computational health information science. They bring methods and concepts of biomedical informatics and data science to bear on problems in basic biomedical and behavioral research, health care, public health, consumer health, and other domains. NLM offers four general categories of extramural support: research project grants; training, fellowships, and career support; information resource awards; and small business grants. NLM also manages relevant awards issued as part of trans-NIH research initiatives, such as the Pioneer Awards. NLM is expanding its extramural programs to support growing demand for innovation in data science.
Biomedical Informatics Research
NLM’s research project grants (RPGs) support pioneering research and development to advance biomedical informatics and data science, including methods for extracting meaning from data, such as genomic sequences or clinical data from EHRs. NLM supports both basic and applied research, ranging from small proof-of-concept projects to large, multi-site research collaborations. In FY 2018, NLM issued 29 new RPGs, including six exploratory/developmental awards. Innovation in data science was a research priority, representing more than 60 percent of funded research grants. In support of the NIH Next Generation Researcher initiative, NLM awarded RPGs to 10 early-stage investigators. Four new awards in translational bioinformatics funded research projects focused on molecular interaction, gene regulation, genetic variants and neuroanatomical shape, and alterations of gene expression patterns. NLM also issued an administrative supplement to the NIH Helping to End Addiction Long-term (HEAL) initiative.
NLM is making increasing use of focused funding announcements to target areas of research important to its mission and broader NIH interests in data science. In FY 2018, for example, NLM launched a new initiative to apply data science approaches to help consumers gather, manage, use, and understand information about their personal health. Five newly-funded research awards in FY 2018 will create tools for patients and caregivers to better manage chronic conditions, including diabetes and epilepsy; self-tracking tools (e.g., activity and symptom sensors) to capture a more complete, accurate, and longer-term understanding of an individual's health and improve self-management and clinical care; and a smartphone application to help motivate and enable African American and Hispanic patients to assemble and use prevention-focused personal health libraries. NLM also launched a new initiative to support computational approaches to curation at-scale. Awards will support novel informatics and data science approaches for increasing the speed and assuring the quality of automated annotation, storage, and retrieval of biomedical research resources (e.g., data, images, models) to make them FAIR.
NLM also established a partnership with the National Science Foundation to support research and training at the intersection of quantitative/computational sciences and the biomedical science.
NLM sets aside funds to support Small Business Innovation Research and Technology Transfer (SBIR/STTR). In FY 2018, NLM funded five new SBIR/STTR awards totaling more than $1.2 million. The new projects allow for improvements to natural language processing for documenting meaningful use of EHRs, block-chain enabled decision support to safeguard privacy and security of patients and research participants, and automated abstraction of information from EHRs.
Informatics Workforce for Biomedical Informatics and Data Science
NLM provides a range of training and career development opportunities in biomedical informatics and data science to create a diverse workforce with skills in developing, applying, and making effective use of emerging data science tools and techniques. Many of today’s informatics researchers and health information technology industry leaders are graduates of NLM-funded university-based training programs in biomedical informatics. NLM’s sixteen active university-based programs offer graduate education and postdoctoral research experience to more than 200 individuals each year in a range of areas, including health care informatics, translational bioinformatics, clinical research informatics, and public health informatics. Supplemental funding and relationships with other NIH ICs extend this influence to cover additional fields of interest and enhance efforts to engage related disciplines and special populations.
In addition to these full-time training opportunities, NLM’s training programs support short-term summer research experiences, which were expanded in FY 2018 to support nearly 40 graduate and undergraduate students. NLM funded six career awards in FY 2018 from two career transition programs that are offered to NLM trainees and others ready to launch informatics research careers. NLM also offers administrative supplements to existing NIH research grantees who want to add an information specialist to their research team. Information specialists help ensure that the research team follows best practices for collecting, managing, and analyzing data in ways that facilitate and support rigorous and reproducible research. NLM plans to extend this successful program in FY 2019 to enhance the ability of other NIH ICs to support their own information specialists as research facilitators.
Taken together, NLM’s commitment to training and career transition in FY 2018 represented more than 30 percent of NLM’s extramural grants budget. NLM intends to continue supporting research training and career transition through institutional training programs, as well as individual fellowships and career grants.
Program Portrait: University-based Research Training program to Support a Diverse, Data-savvy Workforce in Biomedical Informatics and Data Science
FY 2019 Level: $ 12.59 million
FY 2020 Level: $ 11.33 million
Change: -$ 1.26 million
For more than 30 years, NLM’s Extramural Programs Division has been a recognized source of support for research training in biomedical informatics and data science. NLM’s university-based biomedical informatics and data science research training programs offer graduate education and postdoctoral research experiences in a wide range of areas including: health care informatics, translational bioinformatics, clinical research informatics, and public health informatics. In all of these areas, biomedical data science concepts and methods are part of the core curriculum. Seven programs also offer special tracks in environmental exposure informatics.
NLM’s university-based program produces interdisciplinary, cross-trained researchers who understand fundamental principles of knowledge representation, decision support, translational research, human-computer interaction, and social and organizational factors that influence effective discovery and application of new knowledge in biomedical domains. In FY2018, NLM supported institutional training in biomedical informatics and data science at 16 universities across the United States that included some 200 predoctoral and postdoctoral trainees. The National Institute of Environmental Health Services (NIEHS) provided funds to support 14 additional trainees in environmental exposures informatics at seven of the institutions. Two of NLM’s programs received supplemental funds from the NIH Big Data to Knowledge (BD2K) initiative to offer predoctoral training for data scientists.
Each NLM training program is enhancing diversity in the field of biomedical informatics through special recruitment efforts in target populations that are traditionally underrepresented in biomedical research, including Hispanics and African Americans. Cultivating the next generation of leading informaticians and fostering the diverse perspectives that support leading research requires active outreach to underrepresented and disadvantaged groups at earlier educational stages. In FY 2018, NLM offered supplemental funds to support partnerships with minority-serving institutions. Three such supplements were awarded to help facilitate recruitment of high school and undergraduate students into biomedical informatics training programs. To enhance outreach, these supplements emphasized the presentation of student work to bring greater awareness of biomedical informatics and data science literacy to a greater number of diverse young scholars.
NLM has also provided support to bring informatics and data science expertise into other biomedical training programs. Eleven programs used short-term training appointments supported by NLM to provide practicum experiences for prospective trainees. Supplemental funding was also awarded to support partnerships with schools of information, to provide summer research experiences for high school and undergraduate students, and to share curriculum with other NIH-supported training programs.
Resource Grants
NLM sponsors several unique grant programs that support development of biomedical knowledge resources. NLM’s Information Resources to Reduce Health Disparities program develops information resources for underrepresented groups. A new funding announcement was issued for applications to be awarded in FY 2019. NLM’s Grants for Scholarly Works in Biomedicine and Health support preparation of book-length manuscripts of value to U.S. health professionals, public health officials, biomedical researchers and historians of the health sciences. Four new awards were made in FY 2018 on such topics as informed consent in genomic medicine, fluoridation of water, and health impact of poor posture.
Budget Policy:
The FY 2020 President’s Budget estimate includes $55.501 million for NLM’s Extramural Programs, a decrease of $9.110 million, or 14.1 percent, from the FY 2019 Enacted level of $64.61 million. Data science and informatics research is fundamental to the sophisticated information systems used to store, manage, display, and analyze research and health data. NLM will continue to accept investigator-initiated applications through NIH parent-grant announcements, as well as applications submitted to NLM’s own funding announcements, which focus on novel, broadly applicable methods in biomedical informatics and data science, personal health libraries and digital curation. In FY 2020, NLM will apply reductions averaging 17 percent to all noncompeting grants, including resource grants, in order to reduce the impact on new awards. NLM expects to award up to 20 new research project grants and will aim to support early stage and new investigators on at success rates comparable to those of established investigators submitting new applications. NLM will continue to support its unique resource grant programs, career transition programs, at reduced levels. It will continue to support its highly regarded university-based training programs, maintaining the number of university-based training programs.
Research Management and Support (RMS)
RMS activities provide administrative, budgetary, logistical, and scientific support for basic library services, intramural research programs, and the review, award, and monitoring of research grants and training awards. RMS functions also include strategic planning, coordination, and evaluation of NLM’s programs, regulatory compliance, policy development, international coordination, and liaison with other Federal agencies, Congress, and the public. These activities are conducted by the NLM Director and immediate staff, as well as personnel from NLM’s Office of Extramural Programs, Office of Administrative Management, Office of Strategic Initiatives, and Office of Communications and Public Liaison. NLM plans to expand RMS activities related to data science and open science, including coordination of NLM and NIH-wide efforts on health data standards, data curation at-scale, data management, sharing, and access, and data science training, as well as strategically aligning NLM’s internal resources to improve efficiency and leverage common approaches across the Library.
Budget Policy:
The FY 2020 President’s Budget estimate is $17.807 million, a decrease of $1.979 million, or 10.0 percent from the FY 2019 Enacted level of $19.786 million. RMS will support NLM-wide planning and evaluation, including implementation of NLM’s new strategic plan. It will also support enhancement of NLM’s information systems security, policy development and administration functions, and improved coordination of trans-NLM and trans-NIH efforts in data science.
Budget Authority By Object Class 1
(Dollars in Thousands)
| FY 2019 Enacted | FY 2020 President's Budget | FY 2020 +/- FY 2019 |
|
|---|---|---|---|
| Total compensable workyears: | |||
| Full-time equivalent | 741 | 741 | 0 |
| Full-time equivalent of overtime and holiday hours | 1 | 1 | 0 |
| Average ES salary | $184 | $184 | $0 |
| Average GM/GS grade | 11.9 | 11.9 | 0.0 |
| Average GM/GS salary | $106 | $106 | $0 |
| Average salary, grade established by act of July 1, 1944 (42 U.S.C. 207) | $111 | $111 | $0 |
| Average salary of ungraded positions | $148 | $148 | $0 |
| OBJECT CLASSES | FY 2019 Enacted | FY 2020 President's Budget | FY 2020 +/- FY 2019 |
|
| Personnel Compensation | ||||
| 11.1 | Full-Time Permanent | 43,613 | 43,779 | 166 |
| 11.3 | Other Than Full-Time Permanent | 41,708 | 41,867 | 158 |
| 11.5 | Other Personnel Compensation | 1,560 | 1,566 | 6 |
| 11.7 | Military Personnel | 114 | 118 | 4 |
| 11.8 | Special Personnel Services Payments | 1,701 | 1,707 | 6 |
| 11.9 | Subtotal Personnel Compensation | $88,696 | $89,037 | $340 |
| 12.1 | Civilian Personnel Benefits | 27,361 | 27,878 | 517 |
| 12.2 | Military Personnel Benefits | 56 | 58 | 2 |
| 13.0 | Benefits to Former Personnel | 0 | 0 | 0 |
| Subtotal Pay Costs | $116,113 | $116,973 | $859 | |
| 21.0 | Travel & Transportation of Persons | 1,129 | 881 | -248 |
| 22.0 | Transportation of Things | 50 | 39 | -11 |
| 23.1 | Rental Payments to GSA | 0 | 0 | 0 |
| 23.2 | Rental Payments to Others | 198 | 149 | -50 |
| 23.3 | Communications, Utilities & Misc. Charges | 557 | 422 | -134 |
| 24.0 | Printing & Reproduction | 136 | 102 | -34 |
| 25.1 | Consulting Services | 73,224 | 55,710 | -17,514 |
| 25.2 | Other Services | 70,079 | 52,916 | -17,163 |
| 25.3 | Purchase of goods and services from government accounts | 67,818 | 61,074 | -6,743 |
| 25.4 | Operation & Maintenance of Facilities | 1,247 | 938 | -309 |
| 25.5 | R&D Contracts | 3 | 3 | 0 |
| 25.6 | Medical Care | 0 | 0 | 0 |
| 25.7 | Operation & Maintenance of Equipment | 16,762 | 12,708 | -4,053 |
| 25.8 | Subsistence & Support of Persons | 0 | 0 | 0 |
| 25.0 | Subtotal Other Contractual Services | $229,132 | $183,349 | -$45,783 |
| 26.0 | Supplies & Materials | 2,112 | 1,833 | -280 |
| 31.0 | Equipment | 27,955 | 21,212 | -6,743 |
| 32.0 | Land and Structures | 0 | 0 | 0 |
| 33.0 | Investments & Loans | 0 | 0 | 0 |
| 41.0 | Grants, Subsidies & Contributions | 64,613 | 55,503 | -9,110 |
| 42.0 | Insurance Claims & Indemnities | 0 | 0 | 0 |
| 43.0 | Interest & Dividends | 0 | 0 | 0 |
| 44.0 | Refunds | 0 | 0 | 0 |
| Subtotal Non-Pay Costs | $325,884 | $263,490 | -$62,393 | |
| Total Budget Authority by Object Class | $441,997 | $380,463 | -$61,534 | |
1 Includes FTEs whose payroll obligations are supported by the NIH Common Fund.
Salaries and Expenses
Dollars in Thousands
| OBJECT CLASSES | FY 2019 Enacted | FY 2020 President's Budget | FY 2020 +/- FY 2019 |
|---|---|---|---|
| Personnel Compensation | |||
| Full-Time Permanent (11.1) | $43,613 | $43,779 | $166 |
| Other Than Full-Time Permanent (11.3) | 41,708 | 41,867 | 158 |
| Other Personnel Compensation (11.5) | 1,560 | 1,566 | 6 |
| Military Personnel (11.7) | 114 | 118 | 4 |
| Special Personnel Services Payments (11.8) | 1,701 | 1,707 | 6 |
| Subtotal Personnel Compensation (11.9) | $88,696 | $89,037 | $340 |
| Civilian Personnel Benefits (12.1) | $27,361 | $27,878 | $517 |
| Military Personnel Benefits (12.2) | 56 | 58 | 2 |
| Benefits to Former Personnel (13.0) | 0 | 0 | 0 |
| Subtotal Pay Costs | $116,113 | $116,973 | $859 |
| Travel & Transportation of Persons (21.0) | $1,129 | $881 | -$248 |
| Transportation of Things (22.0) | 50 | 39 | -11 |
| Rental Payments to Others (23.2) | 198 | 149 | -50 |
| Communications, Utilities & Misc. Charges (23.3) | 557 | 422 | -134 |
| Printing & Reproduction (24.0) | 136 | 102 | -34 |
| Other Contractual Services: | |||
| Consultant Services (25.1) | 73,224 | 55,710 | -17,514 |
| Other Services (25.2) | 70,079 | 52,916 | -17,163 |
| Purchases from government accounts (25.3) | 57,025 | 50,282 | -6,743 |
| Operation & Maintenance of Facilities (25.4) | 1,247 | 938 | -309 |
| Operation & Maintenance of Equipment (25.7) | 16,762 | 12,708 | -4,053 |
| Subsistence & Support of Persons (25.8) | 0 | 0 | 0 |
| Subtotal Other Contractual Services | $218,337 | $172,554 | -$45,783 |
| Supplies & Materials (26.0) | $2,112 | $1,833 | -$280 |
| Subtotal Non-Pay Costs | $222,520 | $175,980 | -$46,540 |
| Total Administrative Costs | $338,634 | $292,953 | -$45,681 |
Detail of Full-Time Equivalent Employment (FTE)
| OFFICE/DIVISION | FY 2018 Final | FY 2019 Enacted | FY 2020 President's Budget | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Civilian | Military | Total | Civilian | Military | Total | Civilian | Military | Total | |
| Division of Extramural Programs | |||||||||
| Direct: | 18 | - | 18 | 19 | - | 19 | 19 | - | 19 |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 18 | - | 18 | 19 | - | 19 | 19 | - | 19 |
| Division of Library Operations | |||||||||
| Direct: | 250 | - | 250 | 266 | - | 266 | 297 | - | 297 |
| Division of Library Operations | |||||||||
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Division of Library Operations | |||||||||
| Total: | 250 | - | 250 | 266 | - | 266 | 297 | - | 297 |
| Division of Specialized Information Services | |||||||||
| Direct: | 31 | - | 31 | 31 | - | 31 | - | - | - |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 31 | - | 31 | 31 | - | 31 | - | - | - |
| Lister Hill National Center for Biomedical Communications | |||||||||
| Direct: | 48 | - | 48 | 38 | - | 38 | 38 | - | 38 |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 48 | - | 48 | 38 | - | 38 | 38 | - | 38 |
| National Center for Biotechnology Information | |||||||||
| Direct: | 285 | 1 | 286 | 302 | 1 | 303 | 302 | 1 | 303 |
| Reimbursable: | - | - | - | - | - | - | - | - | - |
| Total: | 285 | 1 | 286 | 302 | 1 | 303 | 302 | 1 | 303 |
| Office of the Director/Administration | |||||||||
| Direct: | 53 | - | 53 | 70 | - | 70 | 70 | - | 70 |
| Reimbursable: | 14 | - | 14 | 14 | - | 14 | 14 | - | 14 |
| Total: | 67 | - | 67 | 84 | - | 84 | 84 | - | 84 |
| Total | 699 | 1 | 700 | 740 | 1 | 741 | 740 | 1 | 741 |
Includes FTEs whose payroll obligations are supported by the NIH Common Fund.
| OFFICE/DIVISION | FY 2018 Final | FY 2019 Enacted | FY 2020 President's Budget | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Civilian | Military | Total | Civilian | Military | Total | Civilian | Military | Total | |
| FTEs supported by funds from Cooperative Research and Development Agreements. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| FISCAL YEAR | Average GS Grade |
|---|---|
| 2016 | 11.5 |
| 2017 | 11.5 |
| 2018 | 11.9 |
| 2019 | 11.9 |
| 2020 | 11.9 |
Detail of Positions 1
| GRADE | FY 2018 Final | FY 2019 Enacted | FY 2020 President's Budget |
|---|---|---|---|
| Total, ES Positions | 5 | 5 | 5 |
| Total, ES Salary | 921,901 | 921,901 | 921,901 |
| GM/GS-15 | 23 | 25 | 25 |
| GM/GS-14 | 46 | 49 | 49 |
| GM/GS-13 | 137 | 150 | 150 |
| GS-12 | 113 | 113 | 113 |
| GS-11 | 21 | 34 | 34 |
| GS-10 | 0 | 0 | 0 |
| GS-9 | 12 | 12 | 12 |
| GS-8 | 38 | 38 | 38 |
| GS-7 | 7 | 11 | 11 |
| GS-6 | 2 | 2 | 2 |
| GS-5 | 1 | 1 | 1 |
| GS-4 | 2 | 2 | 2 |
| GS-3 | 3 | 3 | 3 |
| GS-2 | 2 | 2 | 2 |
| GS-1 | 3 | 3 | 3 |
| Subtotal | 410 | 445 | 445 |
| Grades established by Act of July 1, 1944 (42 U.S.C. 207) | 0 | 0 | 0 |
| Assistant Surgeon General | 0 | 0 | 0 |
| Director Grade | 0 | 0 | 0 |
| Senior Grade | 1 | 1 | 1 |
| Full Grade | 0 | 0 | 0 |
| Senior Assistant Grade | 0 | 0 | 0 |
| Assistant Grade | 0 | 0 | 0 |
| Subtotal | 1 | 1 | 1 |
| Ungraded | 274 | 280 | 280 |
| Total permanent positions | 409 | 450 | 450 |
| Total positions, end of year | 700 | 741 | 741 |
| Total full-time equivalent (FTE) employment, end of year | 700 | 741 | 741 |
| Average ES salary | 184,380 | 184,380 | 184,380 |
| Average GM/GS grade | 11.9 | 11.9 | 11.9 |
| Average GM/GS salary | 105,698 | 106,100 | 106,100 |
1 Includes FTEs whose payroll obligations are supported by the NIH Common Fund.
Last Reviewed: February 10, 2026

