
The Pathogen Detection Project and Resources
Pathogen Outbreak Surveillance Partners Foodborne Illness Outbreak Surveillance
Foodborne Illness Outbreak Surveillance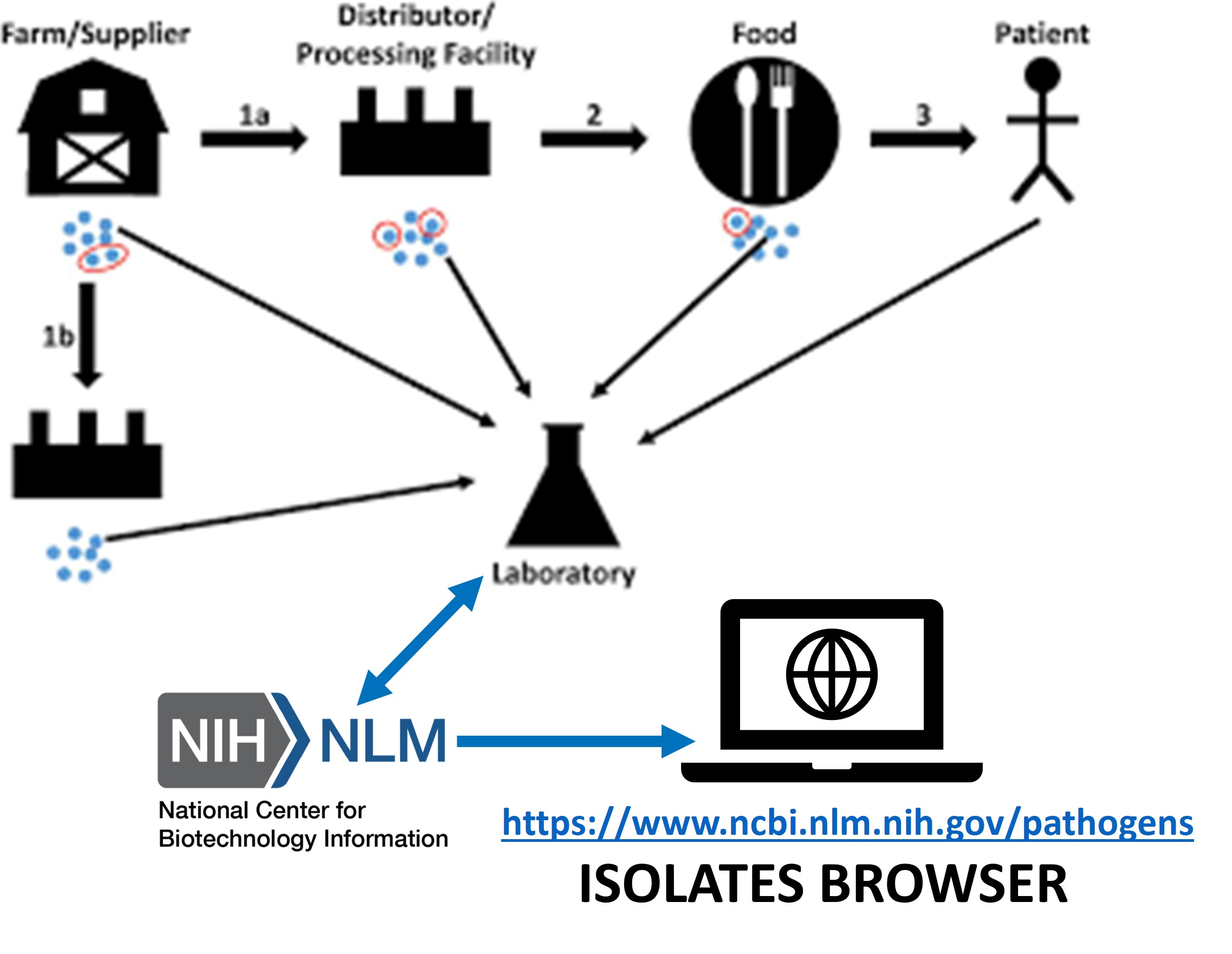
In addition to the RefSeq Project genome curation efforts, the NCBI has other partnerships which are specific in the microbial pathogen community.
- NCBI runs a sequence analysis pipeline for public health laboratory genomic isolates that assembles short-read Illumina sequence data and analyzes it in the context of all other sequences in the system for both genetic relatedness and for AMR surveillance - and then provides a report back to the submitting laboratory.
- To facilitate public health monitoring and anti-microbial resistance research, NCBI also manages and provides access to databases for accessing the isolate information and to identify genes relevant to public health (see below).
- NCBI participates in the consortium of agencies and laboratories for the National Antibiotic Resistance Monitoring System (NARMS) - a unique, publicly available resource that brings together isolate information, AMR phenotypes and genome sequences.
- One of the products is the National Database of Antibiotic Resistant Organisms (NDARO) - a collection of isolates with antibiogram information (measured sensitivity or resistance of a bacterial strain for antibiotic compounds).
To make sure that the research community has access to this critical information, the NCBI Pathogen resource has created these tools:
-
- Isolates Browser - an interactive table that displays data for all available isolates including things such as: identified strain name, isolation type (environmental or clinical) and location, host, and curated antimicrobial resistance, virulence, or stress response genotypes and phenotypes, when available. In addition, they may also have a link to a calculated “SNP Cluster” or other isolates of similar nucleotide sequence.
- Reference Gene Catalog or Hierarchy - interactive tables containing information on key gene families including antimicrobial resistance, biocide and stress resistance, efflux potential, virulence, and antigenicity and links to relevant RefSeq and GenBank nucleotide and protein sequence records.
- Microbial Browser for Identification of Genetic and Genomic Elements (MicroBIGG-E) - an interactive table providing access to genetic and genomic elements with gene family annotations, including those for antimicrobial resistance, stress response or virulence with information about the method and supporting evidence used to identify the element.
-
-
A NCBI Workshop featuring the Pathogen Project's Resources: "Identifying Clinically Relevant Genes in Bacterial Genomes (May 19, 2022)"
NCBI Virus
A team of viral taxonomists and genomics specialists has been working to effectively curate and catalog NCBI viral data for a long time. Recently, they have teamed up a specific development team to produce a community portal to help the scientific community easily find and explore viral sequence data from RefSeq, GenBank and other NCBI repositories.
NCBI Virus is an integrative, interactive resource designed to support retrieval, display and analysis of a curated collection of virus sequences and large sequence datasets. 
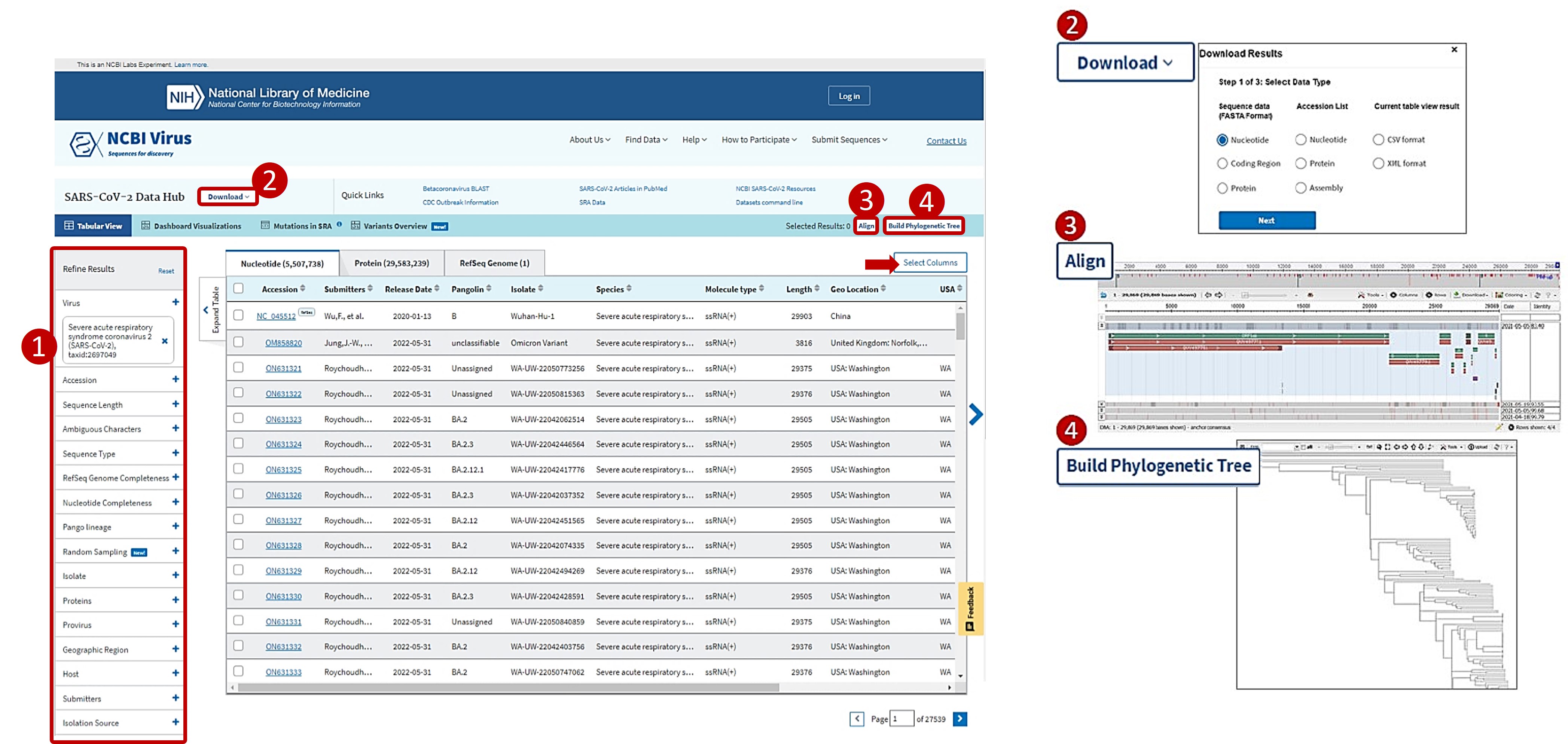
This section isn't very long because, frankly, it is a pretty easy to understand and use resource!
-
- NCBI Minute Webinar recording: "What's new with NCBI Virus?" (June 15, 2022)
Last Reviewed: August 5, 2022

