How BLAST Works
Introduction
We will take a high-level view of the steps performed by BLAST to generate an alignment, with an emphasis on the "words" used to seed BLAST alignments, and we'll briefly discuss Expect values.For more detail, see this explanation of the Blast process.
Global versus local alignments

BLAST overview
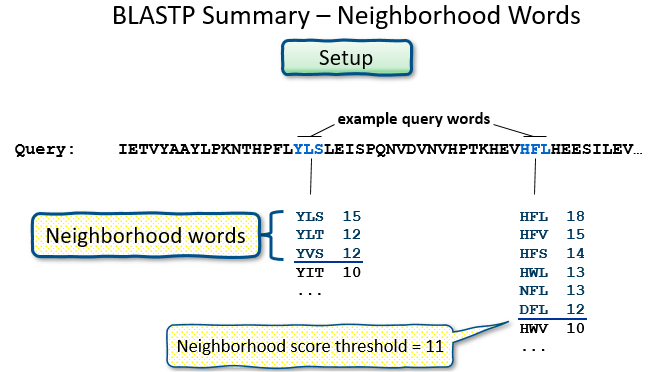
Setup
-
- read in the query, database, and search parameters
- apply query filters, e.g., low complexity and repeats
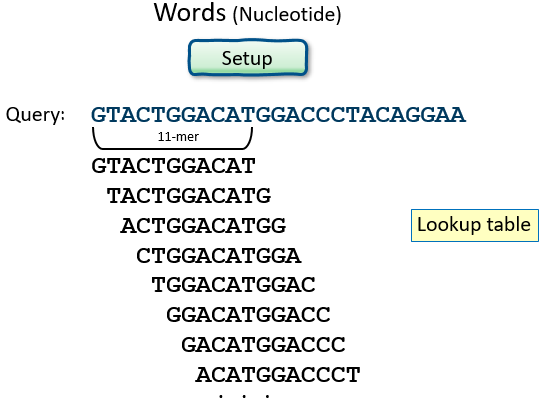
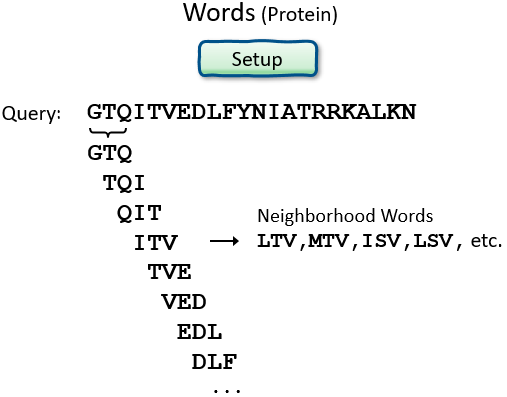
- make a lookup table of query “words”
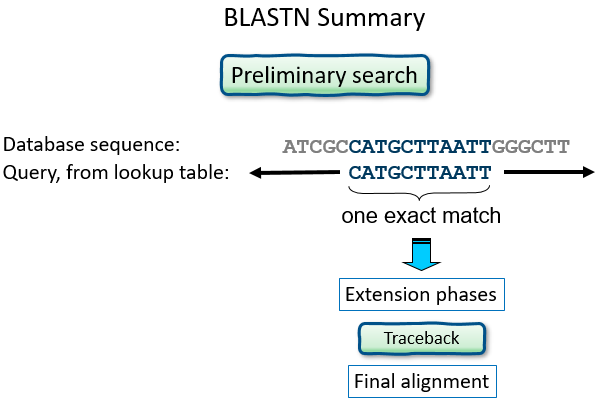
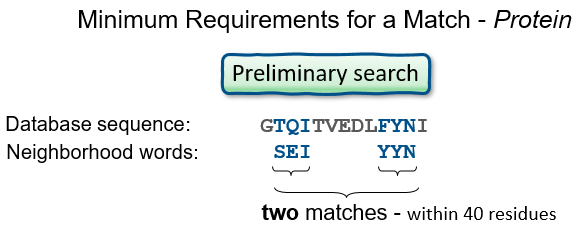
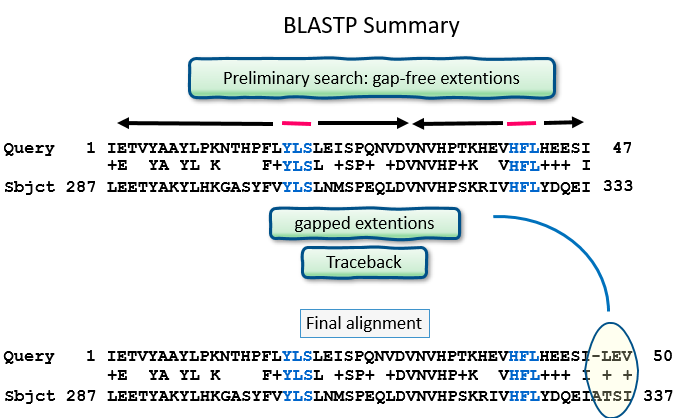
Preliminary search
-
- scan the database for word matches
- gap-free extensions
- gapped extensions, minus deletions/insertions
Traceback
-
-
gapped extensions, calculate the deletions/insertions
-
Nucleotides: Word size, and Summary
Proteins: Word size, and Summary
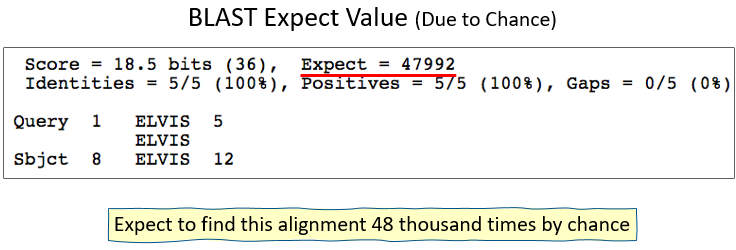
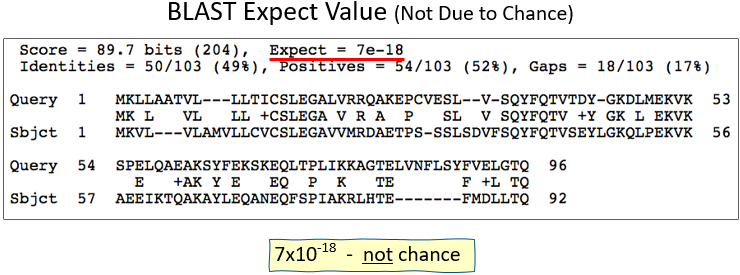
Expect values
| E = number of database hits you expect to find by chance, ≥ S |
Read about: The Statistics of Sequence Similarity Scores
BLAST Expect Value (In a Nutshell)
- E = number of database hits you expect to find by chance
- As the database size increases .... E increases
- As the score increases .... E decreases
Limits, Errors and Warnings
Web BLAST Search Limits
- 5,000 - maximum number of target sequences
- 1,000,000 - maximum sequence length for nucleotide queries
- 100,000 - maximum sequence length for protein queries
BLAST News Feed | NCBI Insights Blog about BLAST settings
Error Messages

Warning Message

Last Reviewed: September 30, 2022

