Demonstration
Below are the high-level topics covered in the demonstration.
These topics closely follow the Help documentation.
The demonstration takes place in CGV.
Topics:
- Set up an alignment
- What are we looking at in the viewer?
- Navigation and zoom
- Gene search
- Link to a view of an alignment in GDV, our Genome Data Viewer
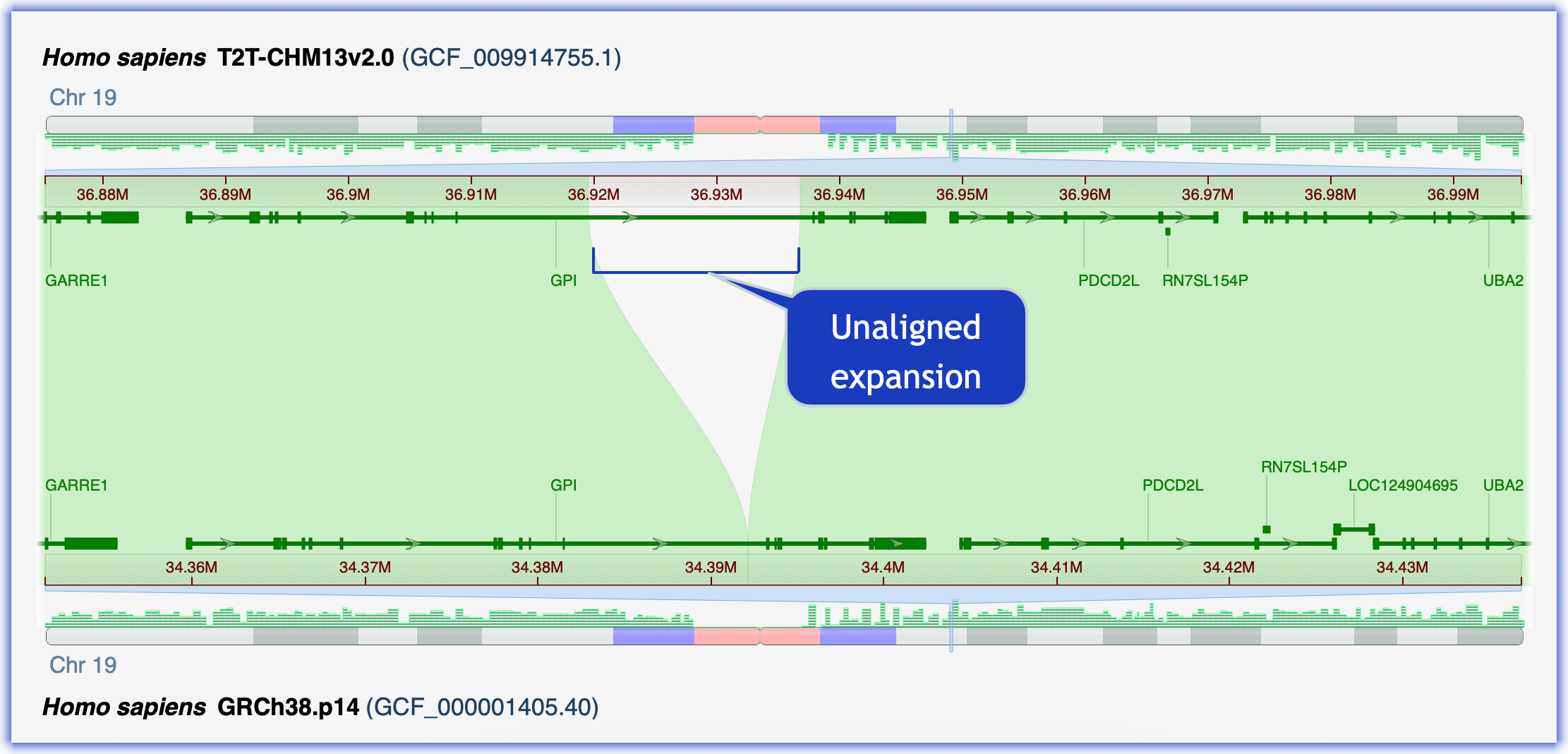
Goal: Analyze an expansion within the GPI gene, glucose 6 phosphate isomerase, in T2T-CHM13v2.0. This region was incorrectly collapsed in GRCh38.p14. GPI deficiency is a rare cause of chronic hemolytic anemia.
Here is the workflow exploring the GPI expansion in T2T-CHM13:
- Set up an alignment between human T2T-CHM13v2.0 and human GRCh38.p14.
- Use "Find a gene in this alignment" to search for glucose 6 phosphate isomerase.
- Click on the gene name under "Description". In this case, either assembly will work.
- Notice the unaligned region in the long intron in the 3-prime half of GPI.
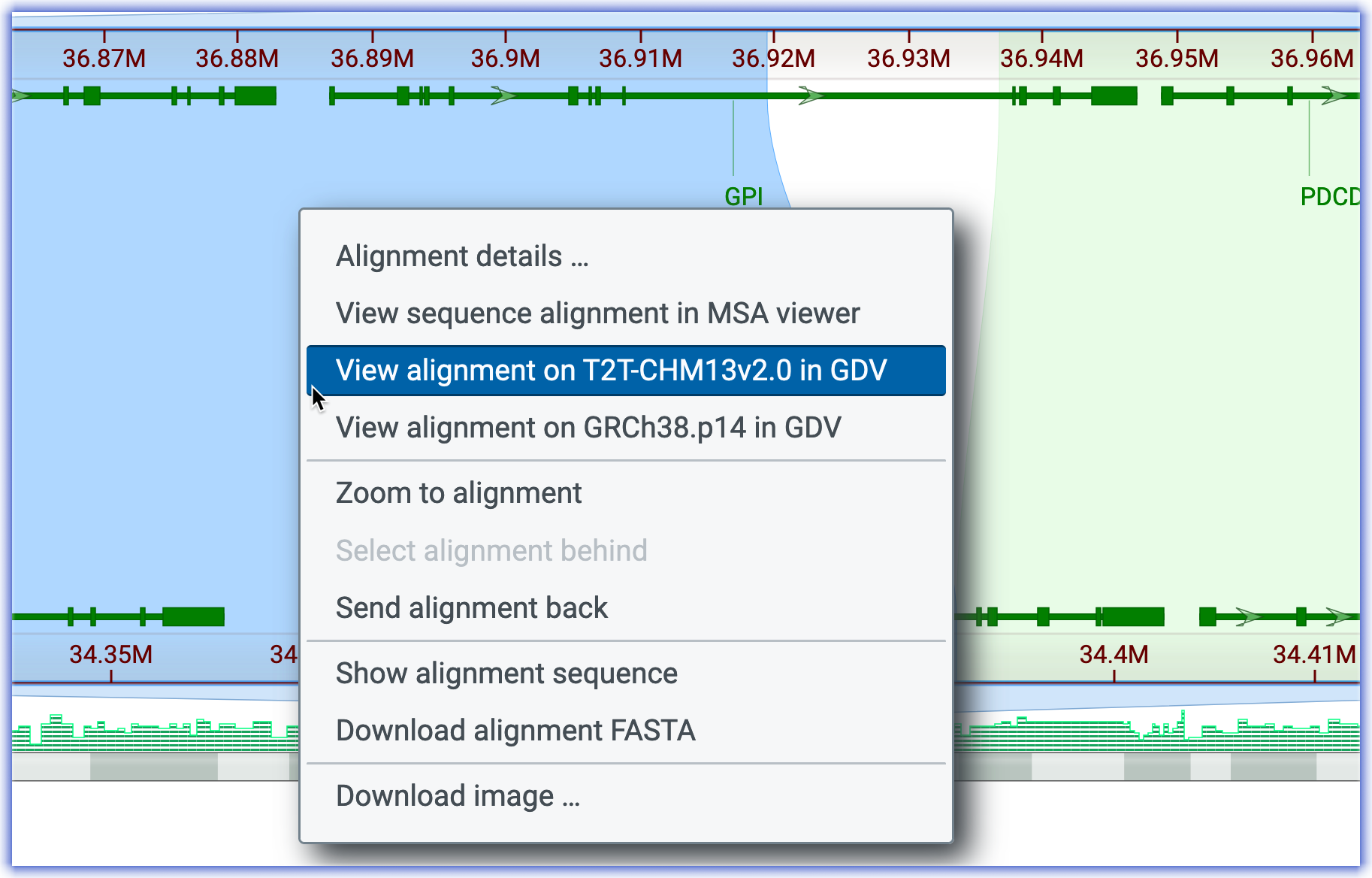
- You can analyze the unaligned region by linking to the Genome Data Viewer, GDV. Right-click on either of the alignments to see the pop-up below. I right-clicked on the upstream alignment, the one on the left. Then select, 'View the alignment on T2T-CHM13v.20 in GDV'
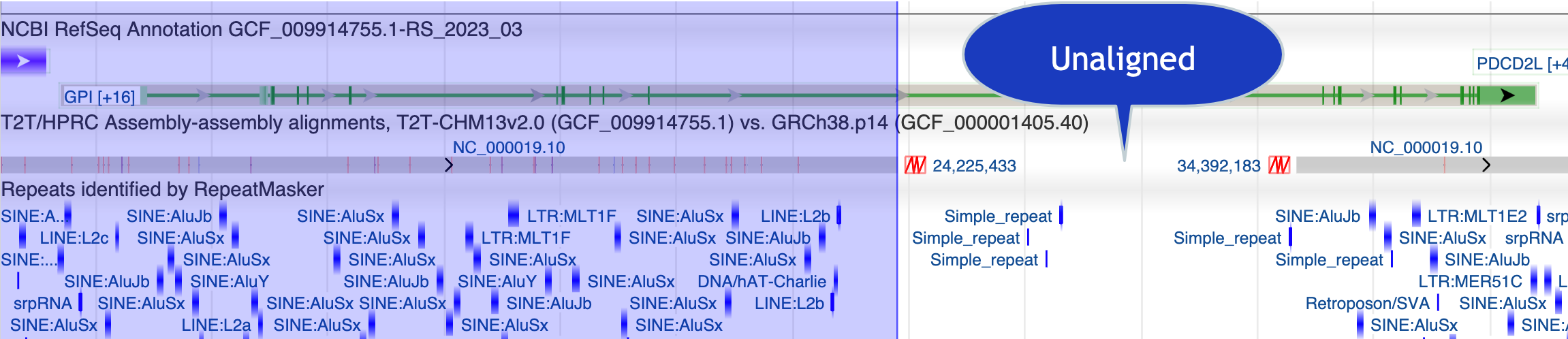
In GDV, if necesary, add the track 'T2T/HPRC Assembly-assembly alignments (T2T aligned to h38)' and the track for 'RepeatMasker Repeats.'
This is one possible view in GDV:
https://www.ncbi.nlm.nih.gov/genome/gdv/browser/genome/?cfg=NCID_1_35266665_130.14.18.128_9146_1688575960_2972660529
That link expires in October, 2023. Here is an image of the view, with an added callout:
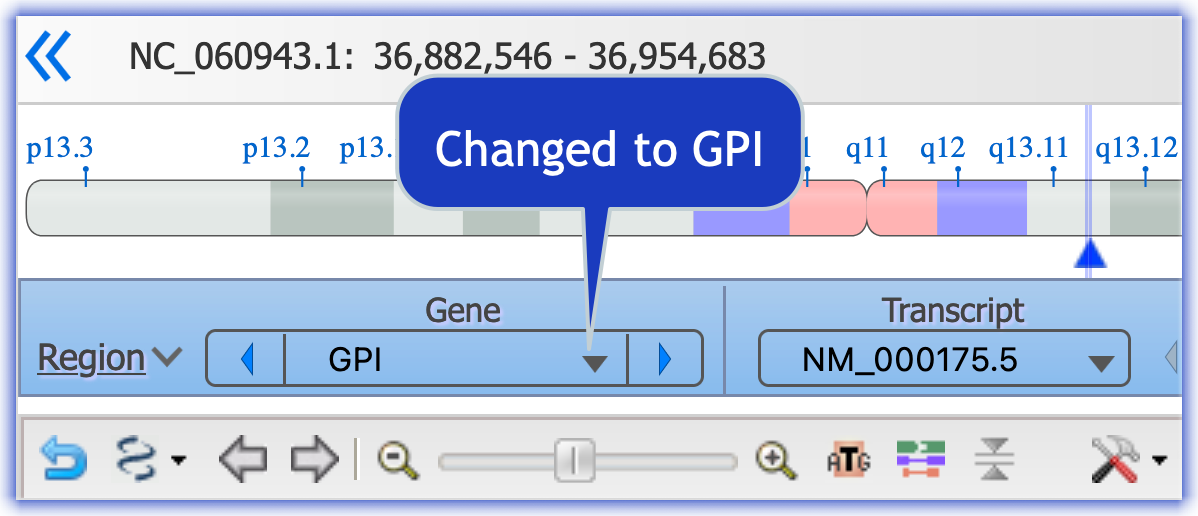
Click here to see the adjustments I made in GDV to get the view above
- Move the track "NCBI RefSeq Annotation ..." to the top
- Use the "Region" menu to focus on the GPI gene
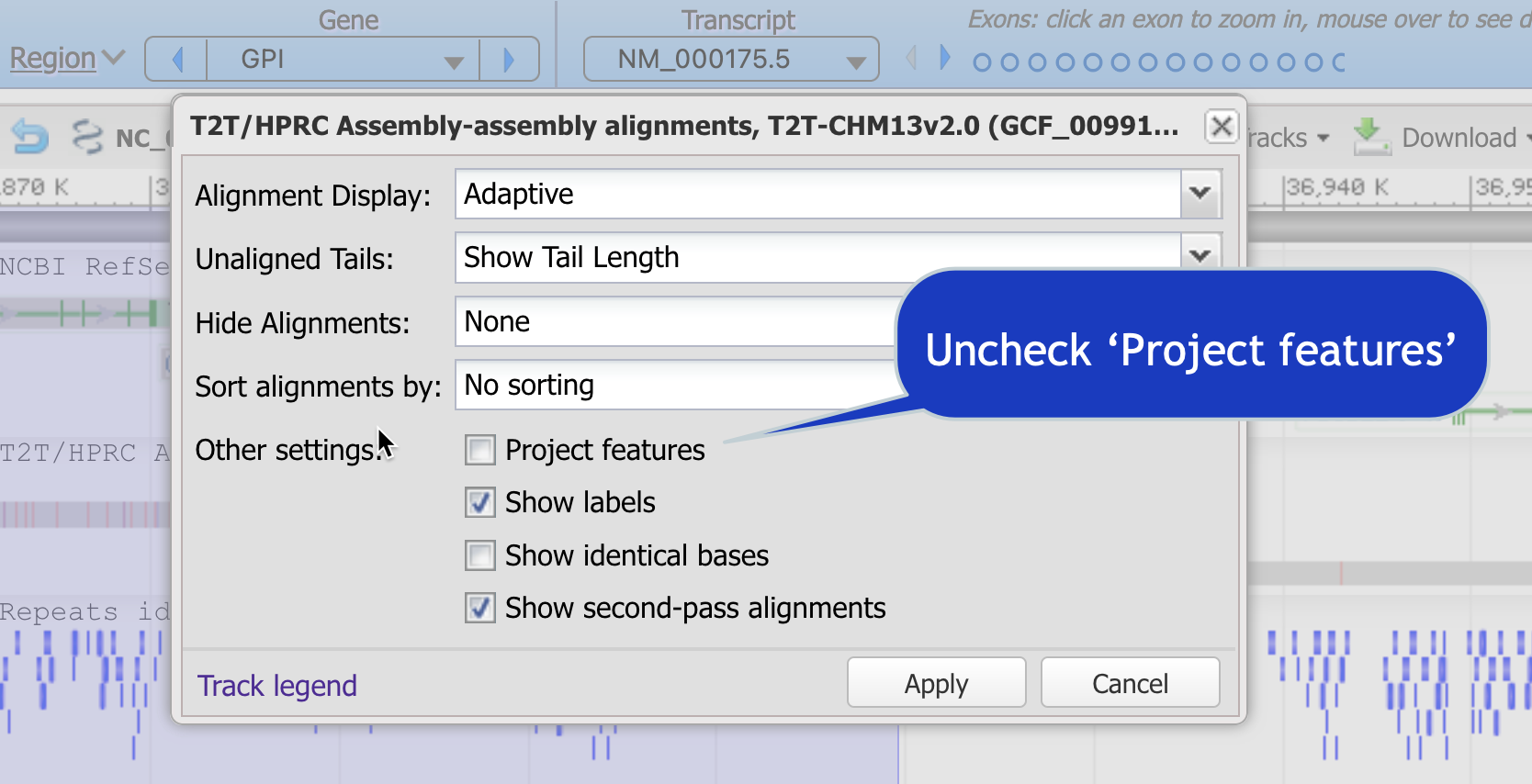
- In the track "T2T/HPRC Assembly-assembly alignments, ..." use the gear icon (Track options) to turn off "Project features." This will collapse the transcripts and proteins for a cleaner view.
Optional Exercise
Identify a repeat unit present in the expanded, unaligned, region in GPI.
Click here to see how to locate a possible repeat unit
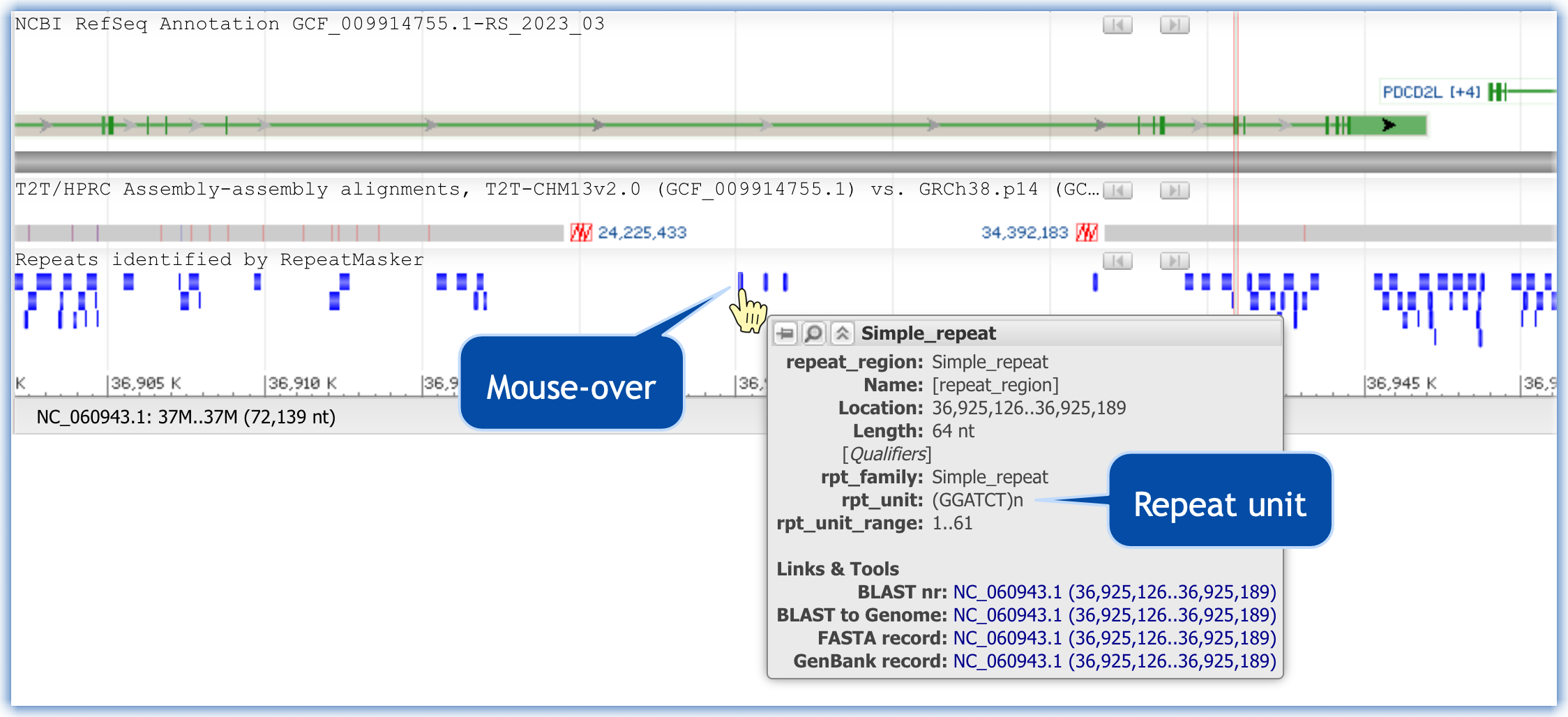
I found more than 400 repeats of GGATCT in the unaligned region of GPI. I downloaded the FASTA for that region to quantify the number of repeats.
Page 1 of 1




