The Primer-BLAST interface
The Primer-BLAST input form has four main sections:
- Sequence input (PCR Template and Primer Parameters)
- Exon / Intron selection
- Primer Pair Specificity Checking Parameters (BLAST database selection and options)
- Advanced Parameters
Input Template or Primer sequences and Primer Parameters
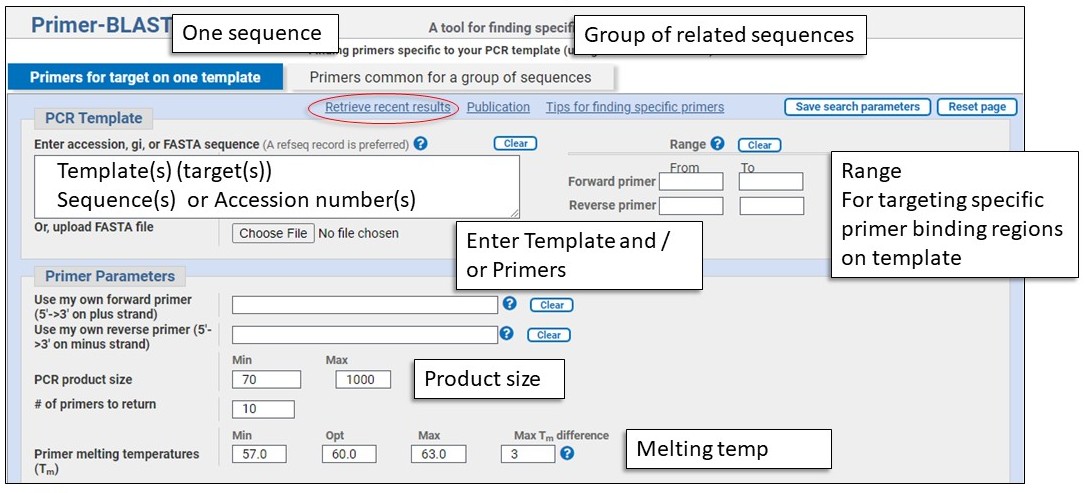
Template and Primers
Design primers:
Combination mode:
Primer parameters
This is where you enter individual forward and/or reverse primer(s), can set the PCR product size, and adjust the Tm parameters if desired.
Exon / Intron Selection
This is where you can set parameters to generate primers specific for mRNA (cDNA) vs. genomic DNA. These settings require an NCBI Reference Sequence transcript (NM_, XM_ accession).
The screen shots below show why requiring that the primers bind to different exons would amplify from the transcript rather than from genomic DNA. Primers binding to exons 7 and 9 of the human myeloperoxidase mRNA (NM_000250) would produce a 750 bp product. The expected product from the genomic region would be over 5 kbp and wouldn't produce any appreciable product under normal PCR conditions.
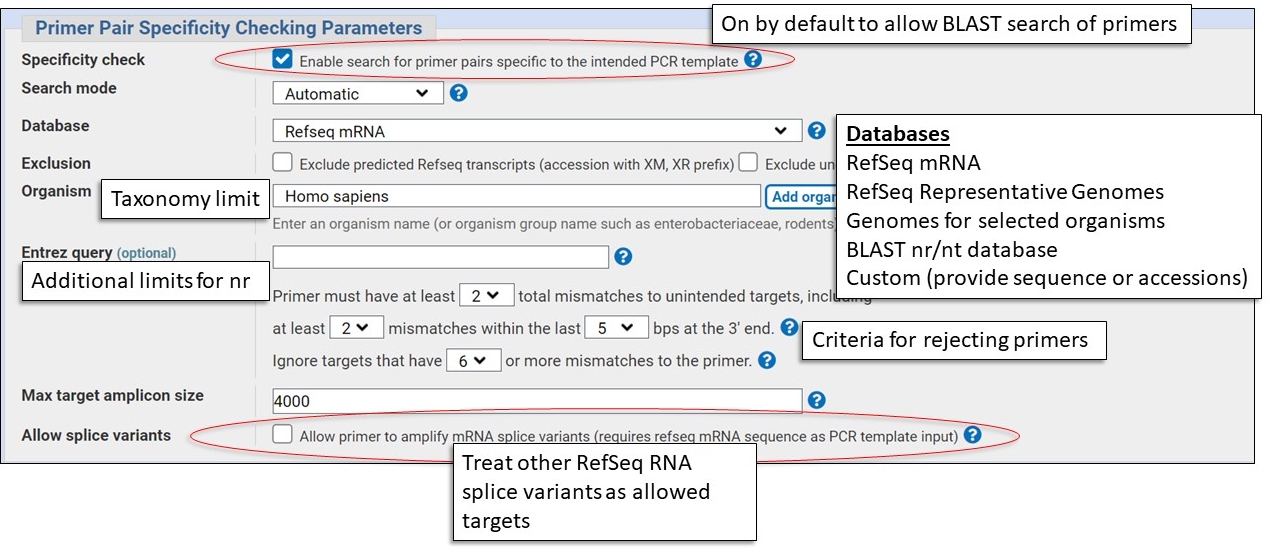
Specificity Checking Parameters
This section is where you select the BLAST database to check for unintended target matches. You should pick the database that best represent the background population of DNA molecules in the sample you'll be using in the PCR experiment, examples: human genomic DNA, human mRNA.
Here are some examples of possible molecular samples and the appropriate database choices. You will nearly always need to specify an organism in addition to a database.
Human cDNA sample
Database: RefSeq mRNA
Organism: Homo sapiens
Human Genomic Sample
Database: RefSeq Representative Genomes
Organism: Homo sapiens
Bacterial Genomic Sample
Database: Representative genomes, nr, custom
Organism: bacterial species or specific strain
Sample from an Organism not in RefSeq
Database: nr
Organism: species or taxonomic group
Note: Use Entrez query to specify genomic or mRNA
biomol_genomic[PROP], biomol_mrna[PROP]
Other Eukaryotic Genomic Sample
Database: Custom, specify the genome assembly accession (e.g., GCA_028021215.1)
Organism: None
Note: Search Genome to find the accession.
Advanced Parameters
Most people rarely adjust the parameters in the Advanced Parmeters. However, if you are using a probe-based quantitative PCR or Real Time PCR, you would request an internal hybridization oligo in this section. You can also request that your primer binding site not contain a known SNP. This only works for human Reference Sequences.
This section also contains the BLAST parameters for the BLAST primer specificity search. The default settings are those for a low stringency BLAST search so that all possible matches are considered. These are similar to the settings triggered when you use a short sequence in a BLAST search. Many people still use BLAST itself as a way to check primer binding targets. In fact, we designed Primer-BLAST in part because we saw that a large number of BLAST searches use short sequences that are likely PCR primers.
Last Reviewed: September 22, 2023