iCn3D Fundamentals
The objective of this exercise is to get familiar with NCBI's iCn3D. We will go over how to navigate, style, color, select, save, and share in iCn3D. The embedded links labeled "follow" are designed to assist participants in the live session with following the instructor's directions. If you fall behind or wish to view a rendering created by the instructor, click these "follow" links.
iCn3D Help Docs
1. Go to iCn3D: Web-based 3D Structure Viewer
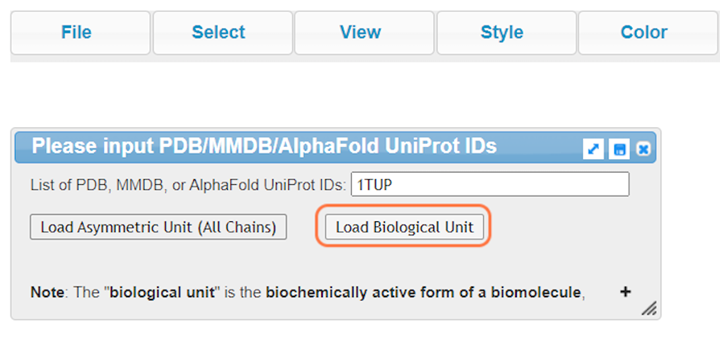
2. Type "1TUP" and Click on Load Biological Unit
3. Orient yourself (see iCn3D Shortcuts and Help Docs)
4. Hover over structure with your mouse to view residues
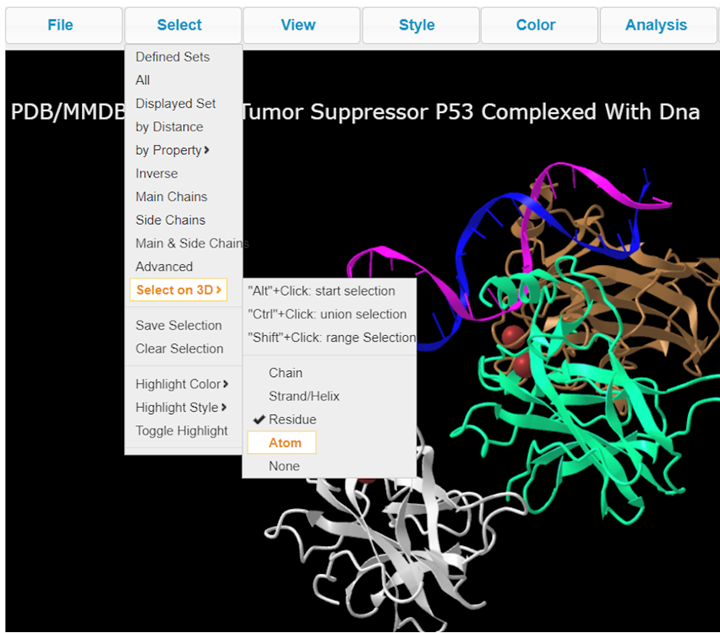
5. Select > Select on 3D > Atom to see atomistic details
6. Hover over structure with your mouse to view atoms
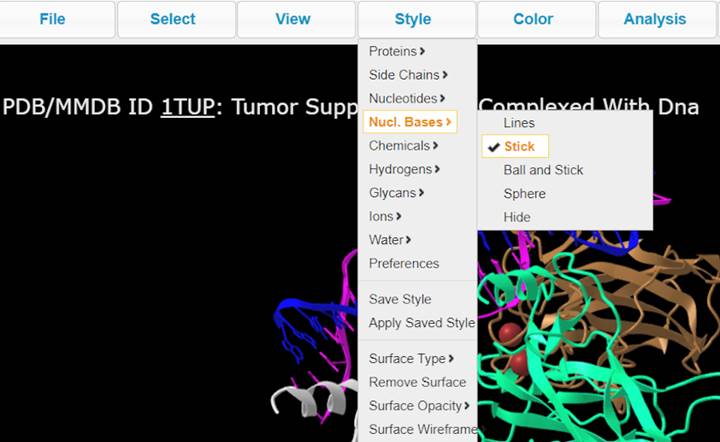
7. Explore different Style options and color the biomolecule to your liking (ex. Nucl. Bases > Stick, Side Chains > Stick)
Follow
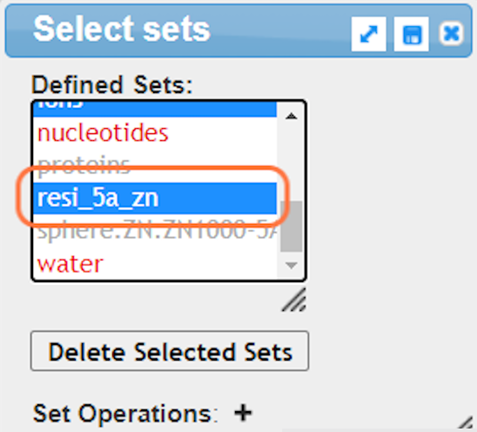
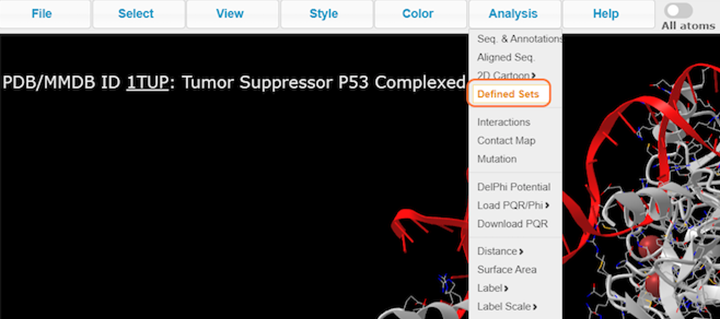
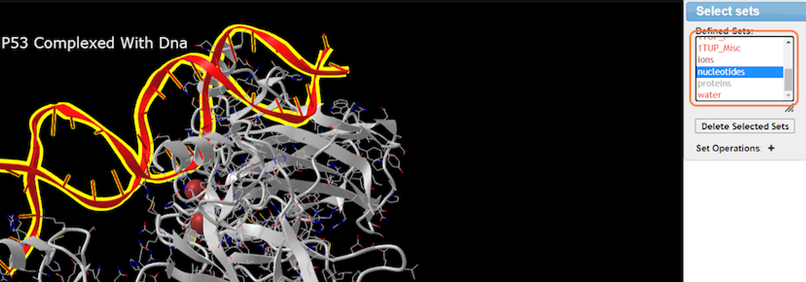
8. Make specific selections with Analysis > Defined Sets
9. Select nucleotides and change color with Color > Rainbow
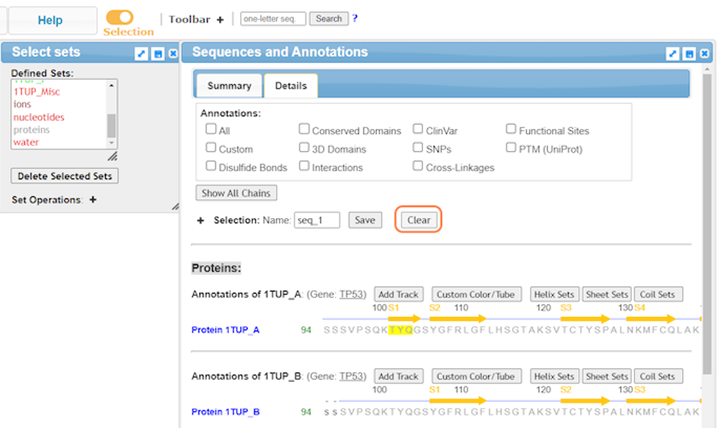
10. Select residues by sequence with Analysis > Seq. & Annotations
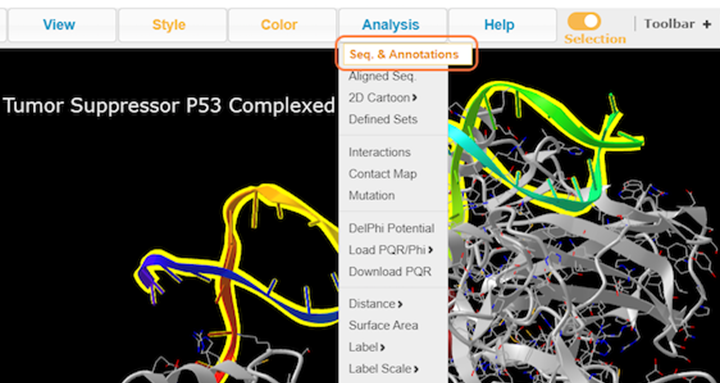
11. Explore the Sequence and Annotations window
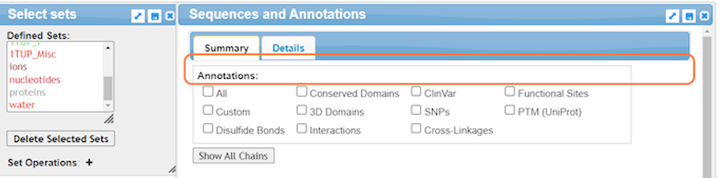
This feature allows you to interactively view and interpret structural features of the biomolecule in the context of sequence information, review functional sites residues, explore the linked data sources, and more! Some guidance on the type of information available:
- Conserved Domains: Highlight regions essential for protein function that haven't changed much over time.
- ClinVar: Show mutations linked to human diseases, helping interpret their potential impact.
- Functional Sites: Pinpoint crucial regions for protein activity, like binding pockets or enzyme active sites.
- Custom: Allows you to add your own data or annotations for personalized analysis, exploration, and hypothesis testing on your biomolecule's sequence and structure.
- 3D Domains: Reveal independent structural units within the protein, offering insights into folding and function.
- SNPs: Show single nucleotide polymorphisms, providing genetic variation data.
- PTM (UniProt): Highlight post-translational modifications like phosphorylation, affecting protein function.
- Disulfide Bonds: Identify covalent bonds between cysteine residues, stabilizing protein structure.
- Interactions: Map interactions between the protein and other molecules, like DNA or protein partners.
- Cross-Linkages: Visualize chemical connections between protein residues, providing structural restraints.
- Transmembrane: Identify regions spanning the cell membrane, essential for membrane proteins' function.
12. For a simplified view, uncheck annotations and click Details
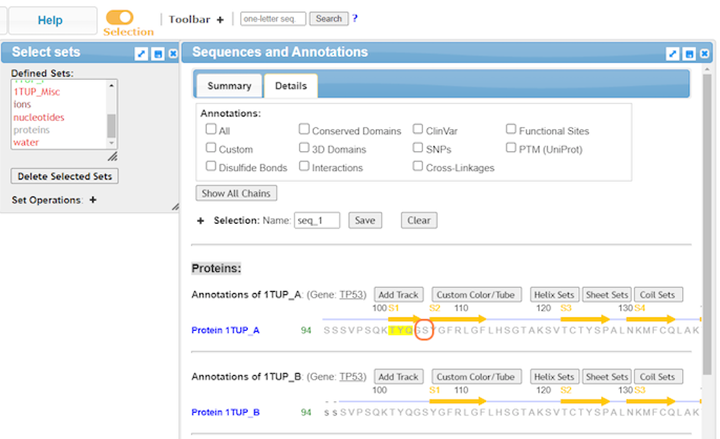
13. Highlight residues from the sequence to select
14. Clear selections and close or minimize Seq. & Annotations when not using
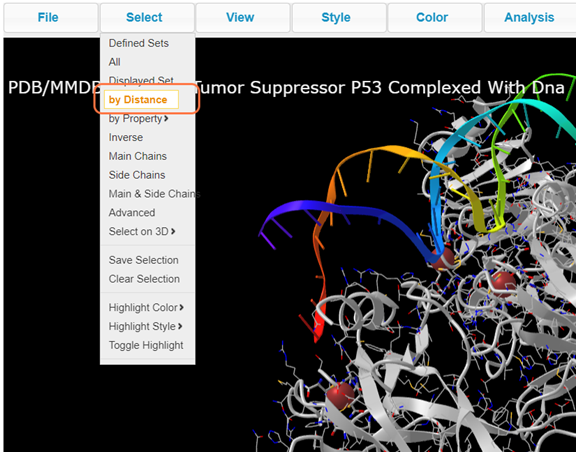
15. Explore ion interactions with Select > By Distance
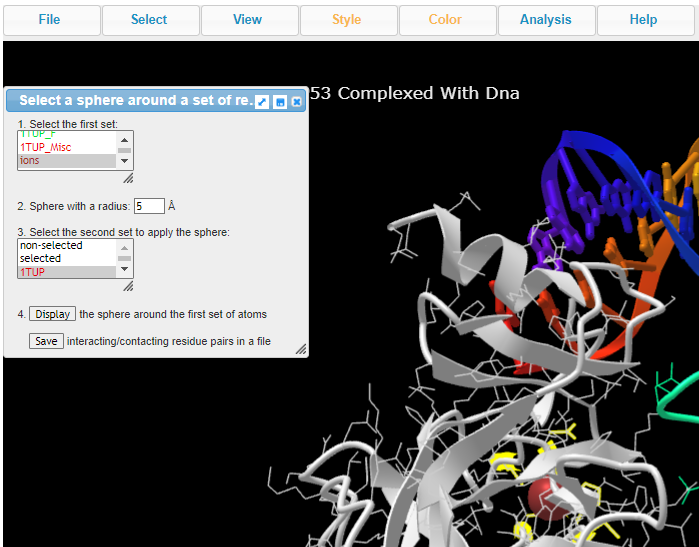
16. Choose ions, set sphere radius to 5 Å, choose 1TUP, and click on Display
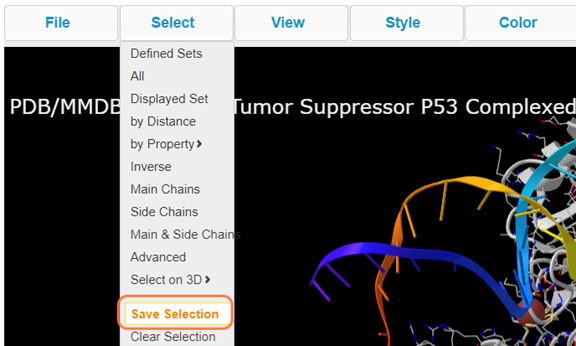
17. Save the selection by Select > Save Selection and give name like resi_5a_zn
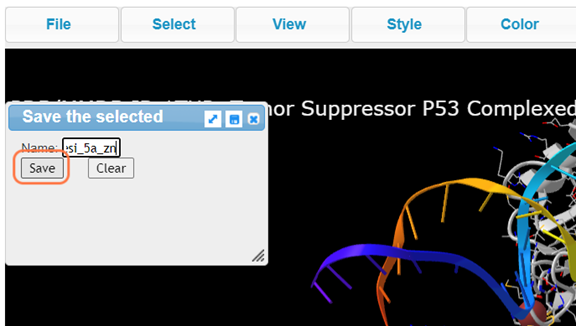
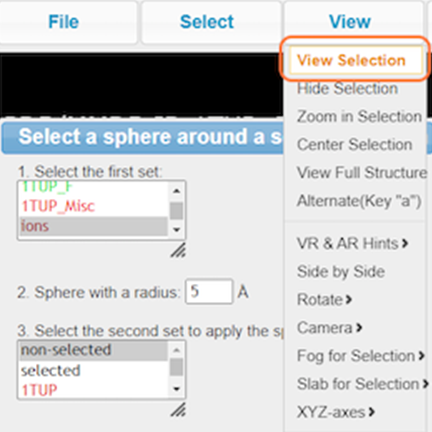
18. View your newly named selection resi_5a_zn and View > View Selection and render to your liking (ex. Style > Side Chains > Stick)
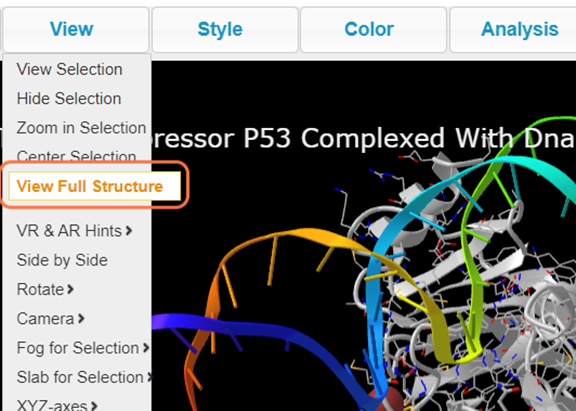
19. Click on View Full Structure to see entire biomolecule again
Follow
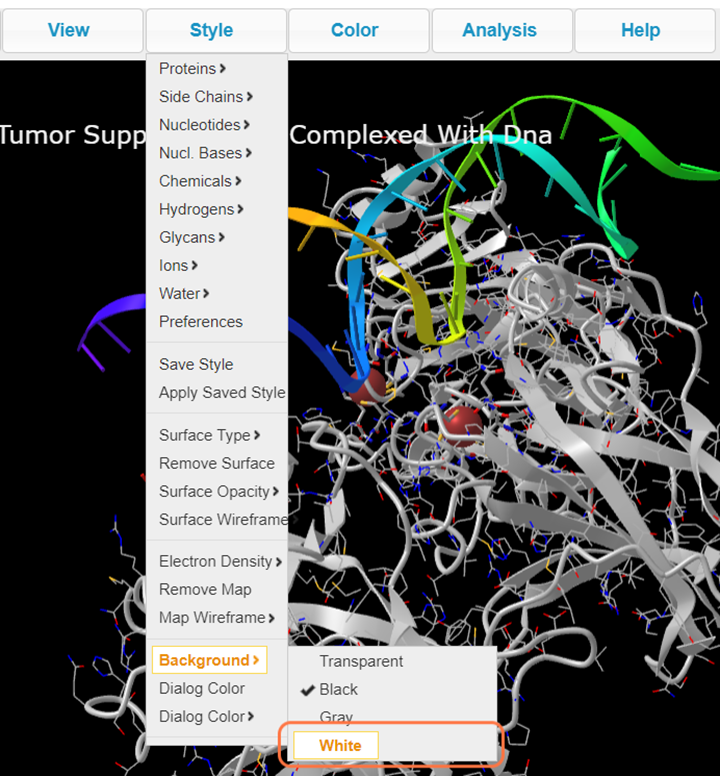
20. Before saving, change your background color with Style > Background > White
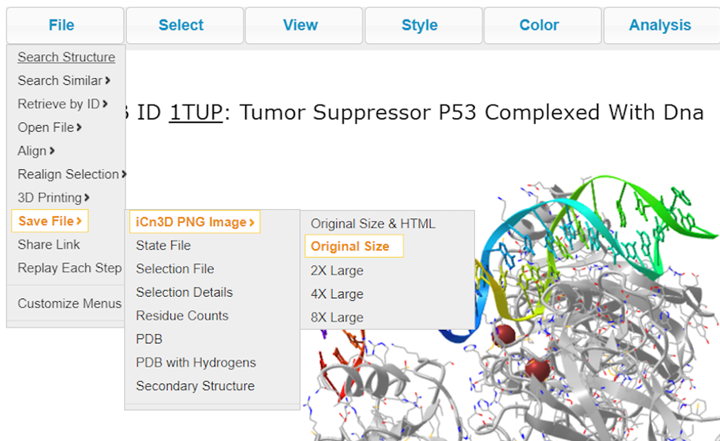
21. Save your rendering as a PNG with File > Save Files > iCn3D PNG image
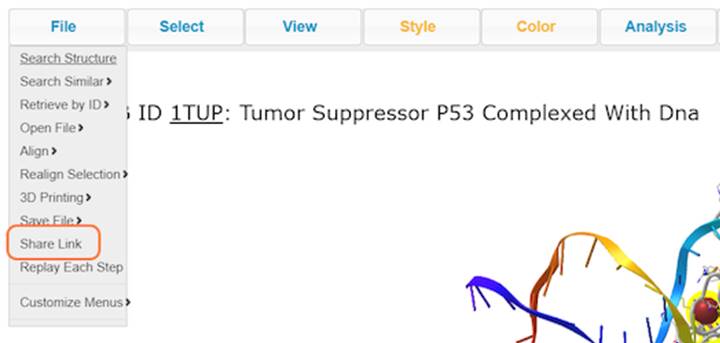
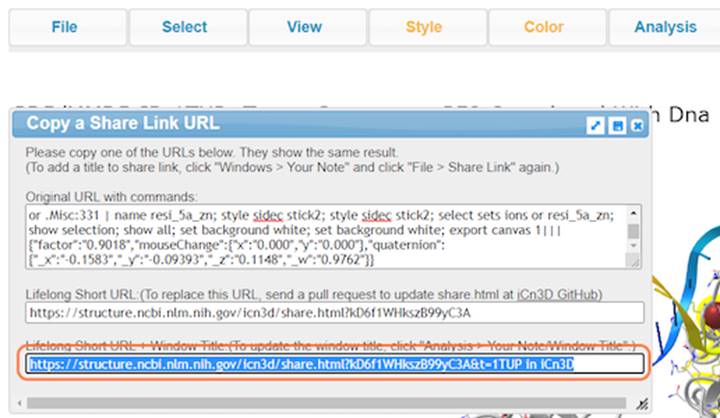
22. Save your rendering as an interactive link with File > Share Link and copy
Practice
Render a biomolecule of interest. If you select something accidentally Select > Clear Selection. If you need to undo View > Undo. Get additional help by 1) Show Help > Help Docs, 2) Help > Selection Hints, or 3) ask us!
Next steps
We will now split into groups to work on one of the three examples:
Example 1: TP53 Mutation Analysis
Example 2: TP53 from Structure to Function
Example 3: Compare Crystal and AlphaFold TP53 Structures
Review the objectives on each of these pages and decide on an example that best suits your learning goals. Work with your group and NCBI experts on completing the steps and rendering an image(s) with iCn3D.
Last Reviewed: June 7, 2024