Clinical Toolkit: Genetics Add-on & NCBI research tips
How can knowing about a patient's genetic variation help my patient care and case management?
- Validate and specify a precision diagnosis
- A patient's specific variant can be used to diagnose a condition or identify a subtype based on the specific molecular lesion
- Understanding the precide variant impact may be able to target preventative and/or monitoring efforts even before clinical features become evident
- When appropriate (such as in the chronic or acute phase of a disorder or condition), the specific molecular lesion may help to precisely select an effectively targeted therapy.
- Aid in customizing a therapeutic outcome
- A patient's known variant may help to uncover an unexpected issue with a prescribed medication's effectiveness due to imact on one or more ADMET issues (absorption, distribution, metabolism, excretion & targeting processes)
- Knowing about the existance of a critical variant in a patient may help to customize effective drug selection and optimal dosage.
Vocabulary: Variant types
When understanding how variants arise or are described, particularly in the clinical community - there are two classes of genetic variants and source tissue samples
- Inherited (germ-line) variants - can be identified in samples from cheek or nasal swab, sputum or blood sample
- Acquired (somatic) variants - would be identified in specific suspect tissue samples (ex: biopsy)
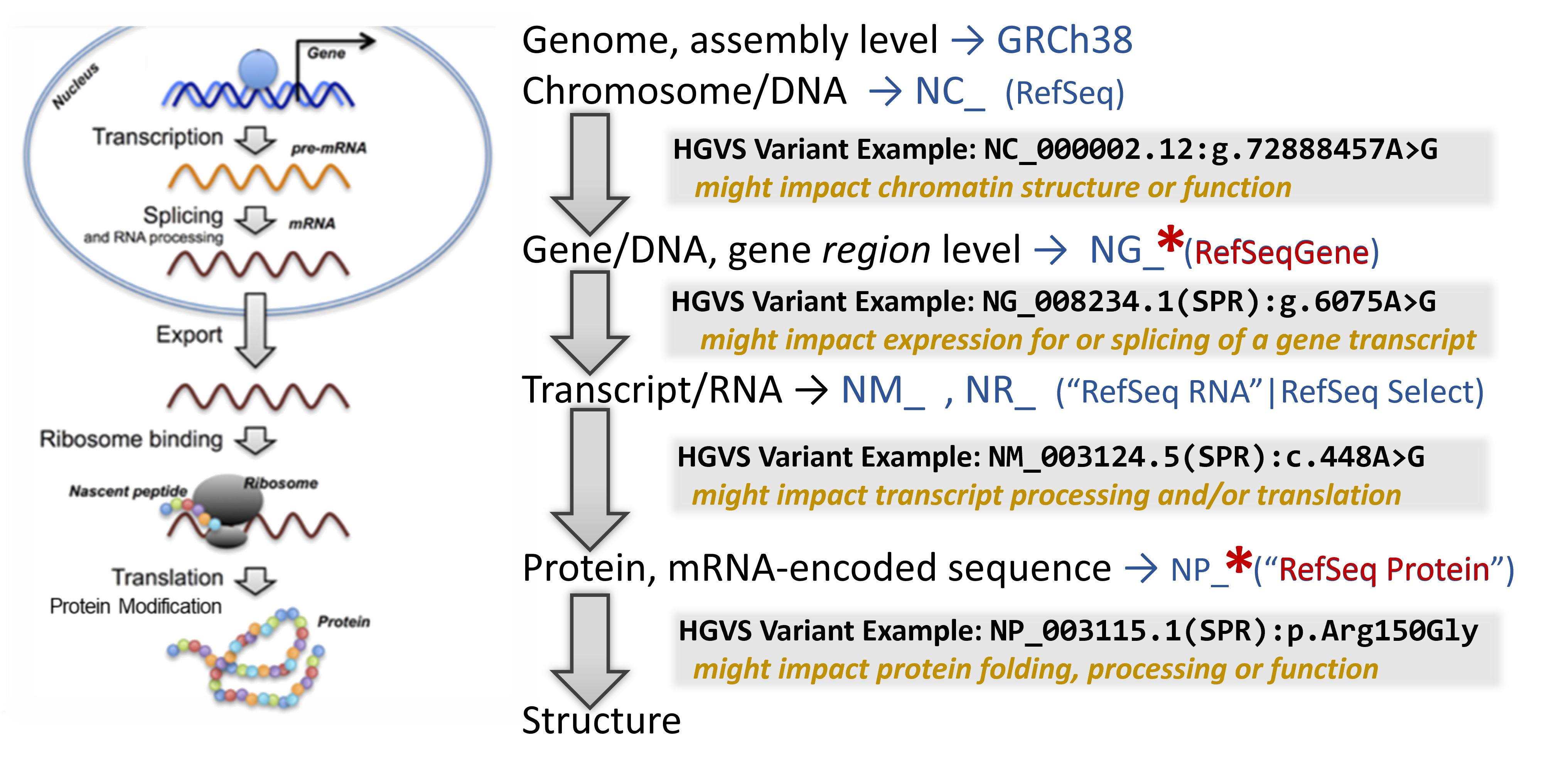
A quick primer on the central dogma & how genetic variants can impact molecular biology
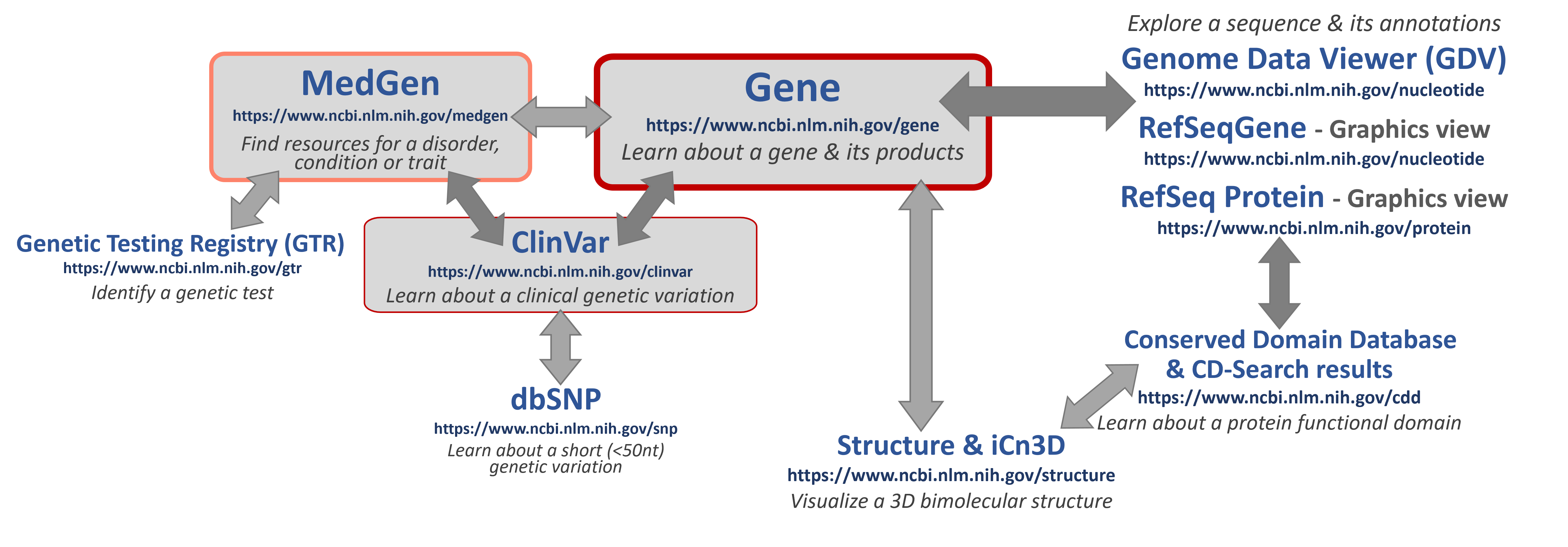
Some helpful NCBI resources
If you are looking for specific information, click here to learn about some good places to START to learn about key topics!
-
- MedGen - an aggregate database for information and links to more about gene-associated human disorders and phenotypes.
- Genetic Testing Registry (GTR) - a source for clinical and research genetic tests with information provided by the laboratories.
- ClinVar - a genetic variation resource collating clinically-relevant information submitted by research and clinical labs, expert clinical panels, and some key genetic disease literature resources.
- NCBI Gene - an aggregate database for information and links to more data for genes
- Nucleotide & Protein - biomolecular sequence databases which include original laboratory-generated, annotated and submitted sequences (such as those in GenBank) as well as more curated, high-quality, annotated sequences (such as those created by the NCBI RefSeq project). The Graphics View enables interactive visualization and exploration of these sequences. For eukaryotic sequences (such as human), the Genome Data Viewer (GDV) will enable you to explore entire or regions of chromosomes.
- NCBI Structure - a source for curated 3D biomolecular structure information based on submissions to the Protein DataBank (PDB). The iCn3D tool is available to interact with and see what they look like!
Vocabulary: Variant nomenclature - How can we notate the presence of a variant?
Over the years, researchers have adopted many different ways to name a particular genetic variant that they have been studying. Here are some examples of what has been used in published literature for exactly the same genetic variant:
-
-
- Factor V Leiden variant
- F5 Arg534Gln
- FV R506Q
- NC_000001.10:g.169519049=
- NC_000001.11:g.169549811C>T
- rs6025
- OMIM: 612309.0001
-
A standard way to notate these has been proposed - the Human Genome Variant Syntax (HGVS) and is now often in use by many research and clinical labs. This notation is based on anchoring the location of a variant to a specific, discrete sequence record with inclusion of a sequence record accession. It is common to include a gene symbol in the syntax, as knowing what gene or protein the accession refers to is not intuitive. Though this practice (as shown below) is recommended, it it not always included and appears to officially be optional.
Accession.version(gene symbol):molecular type-abbreviation.
then a structured statement including:
variant-location, wildtype-residue and variant-impact (such as the variant residue)
For example:
NG_011806.1(F5):g.41721G>A or NP_000121.2(F5):p.Arg534Gln
To learn more about components of HGVS, click here!
For Accession.version, a particular set of sequences and their accessions is often used to reliably and sustainably anchor these variants. The NCBI RefSeq Project collects all known nucleotide and protein sequences and uses them and literature information to create a non-redundant set of reference sequences (recognizable accession prefixes). For humans, these are the most commonly found record-types and prefixes for their accessions:
-
- Chromosome: NC_
- Gene/Gene region: NG_
- Transcript
- Protein coding: NM_ (with strong evidence) or XM_ (predicted)
- Non-protein coding: NR_ (with strong evidence) or XR_ (predicted)
- Protein with sequence translated from the transcript: NP_ (with strong evidence) or XP_ (predicted)
If included, the gene symbol used should be the official one as designated by the Human Genome Nomenclature Committee (HGNC).
For molecular type-abbreviation, at NCBI you will often see:
-
- “g.” for a linear genomic reference sequence
- “c.” for a coding DNA reference sequence
- “p.” for a protein reference sequence
For more information on specific nomeclature for residue designations and indicators (ex: >), please see the documentaion on the the Human Genome Variant Syntax (HGVS) website.
Vocabulary: Genetic testing methods - How can we identify a genetic variant?
Reporting of variant information is often based on the test ordered and identified clinical phenotype, if included.
Targeted Variant tests
-
- Variants reported: Specific, pre-designated variants based on a designated clinical phenotype
-
- Common methods:
- Hybridization/Array
- (increasingly) Next-generation Sequencing (NGS)
- Common methods:
Variant Scanning/Sequencing tests
-
- Variants reported: Laboratories will report on relevant variants if a clinical phenotype code (ICD-10, SnomedCT, HPO ID, OMIM ID) is provided, as is preferred. However, if a clinical code is not specified and one cannot be obtained - all identified with a specific classification (based on the laboratory's policy) may be reported.
- NOTE: Sometimes during NGS analysis a pathogenic or likely pathogenic variant may be detected for a disorder other than indicated by the provided clinical code. This is considered a "Secondary Finding" and may be reported to the ordering clinician. The ACMG makes recommendations for reporting these (see below).
- Variants reported: Laboratories will report on relevant variants if a clinical phenotype code (ICD-10, SnomedCT, HPO ID, OMIM ID) is provided, as is preferred. However, if a clinical code is not specified and one cannot be obtained - all identified with a specific classification (based on the laboratory's policy) may be reported.
-
-
Common methods: Next-generation Sequencing (NGS) or Long-read Sequencing
-
Single Gene Test (single genome region)
-
Multi-gene Panel (several genome regions)
-
mDNA test (mitochondrial genome sequence)
-
Exome Sequencing (transcript sequence set)
-
Whole Genome Sequencing (complete nuclear chromosome set)
-
-
Vocabulary: Clinical variant classifications - How is the phenotype of a variant reported in genetic test results?
| Currently used diagnosis classifications | Under consideration for additional diagnosis classifications | Currently used pharmacogenetic classifications |
Disorder-causing variants
*An important term to know: |
ACMG: Predisposing variants - Those that increase an individual’s susceptibility to a certain disorder or condition, but where the development of symptoms are not certain (used with the qualifier pathogenic or likely pathognic). ClinGen:
|
Drug response variants
Note: The above were based on CYP450 isoform variants. Some additional terms may be released soon by CPIC. |
Some key organizations who provide guidance with regard to clinical genetic variants
| Disorder diagnostic variations | ||
| The American College of Medical Genetics and Genomics (ACMG) - supports efforts to improve patient care, establishes standards of care and laboratory policies, and educates members about advances important to their practices. | ClinGen - an NIH-funded collaborative effort to define the clinical relevance of genes and variants for use in precision medicine and research | Disorder-specific organizations often support panels of experts in a particular topic to produce reference information for use by clinical practice groups, such as Malignant Hyperthermia Associations: MHAUS and EMHG |
| Drug selection and dosage variations | ||
| The Clinical Pharmcogenetics Implementation Consortium (CPIC) - curates and posts, peer-reviewed, evidence-based, updatable, and detailed gene/drug clinical practice guidelines. | The Pharmacogenomics Knowledgebase (PharmGKB) - a resource that aggregates Pharmacogenomics Research Network (PGRN)-curated information about the impact of genetic variation on drug response for clinicians and researchers. | The U.S. Food and Drug Administration (FDA) - maintains a table of drug-gene interactions with sufficient scientific evidence to suggest that subgroups of patients may be likely to have altered drug metabolism. FDA approved guidelines for incorporating genetics in selection or dosage for a specific medication are listed on relevant drug labels. |
Want to learn more about applying a person's genetics in the clinic?
Let's put this all together in a general workflow that you can use
Last Reviewed: August 22, 2023





