Exercise 2: Learn about the pathogen's genome, download and visualize genomic data
To perform helpful bioinformatic analysis for your research it is imperative to find and download high quality sequence data. NCBI accepts nucleotide sequence data from research labs all over the world, but we also have a curation group who uses this data as well as published information to create high quality reference sequences and datasets for use in computational work.
You will perform a search to find a genome record for the pathogen, learn more about the genomic assembly, sequence and annotation, how to visualize and explore this data, as well as how to download the genome, transcriptomes and proteome sequences for further research.
Background
- NCBI: GenBank | Sequence Read Archive (SRA)
- EMBL-EBI: European Nucleotide Archive (ENA) | ENA Read
- Japan's NIG: DNA Data Base of Japan (DDBJ) | DDBJ Sequence Read Archive (DRA)
- The databases contain records of varying quality for both their metadata (descriptive information) and the sequences.
- They also contain sometimes highly redundant sets of data provided independently from a few to hundreds of research labs.
NCBI has developed the Reference Sequences (RefSeq) Project (https://www.ncbi.nlm.nih.gov/refseq/) created/curates high-quality nucleotide and protein sequences from submitted data supplemented with information from peer-reviewed, published literature. The data is produced and therefore “owned” and updated by NCBI.
The project aims to:
- Records represent all molecules in the central dogma
- Provide reference standards
- Eukaryotes: genomic, mRNA & ncRNA, proteins
- Prokaryotes and Viruses: genomic, ncRNA & protein (no mRNA records)
- genomic: NC_, AC_, NG_, NZ_
- RNA: NM_, NR_, XM_, XR_
- protein: NP_ (YP_), XP_, *WP_
*NOTE on on the WP accession.
A Solution: Make one copy of a “shared protein sequence” to link all annotations in the Identical Proteins Group database.
For example: The gene carbapenem-hydrolyzing class A beta-lactamase is annotated on more than 4,700 Bacterial genomic assemblies. It's encoded protein is included in the Identical Proteins Group (IPG) database and report with the accession: WP_004199234.1, MULTISPECIES Taxonomic Group carbapenem-hydrolyzing class A beta-lactamase KPC-2 [Bacteria]
What are genomic assemblies and what can you find at NCBI?
The genome of an organism consists of a set of one or more chromosomes. NCBI's Genome Assembly Model supports a single "assembly" record which lists metadata about the genomic construct as well as a collection of links to individual chromosome sequence records. Assemblies available at NCBI are either created by the research community and submitted to GenBank (assigned accessions beginning with GCA_) or created/curated with in-house annotation pipelines by the NCBI RefSeq Project Team (with accessions beginning with GCF_).
Currently, NCBI has almost 1.8 million genome assemblies for over 144,500 species. For your own organism-of-interest, which one might you prioritize if you need to focus on one to start?
Prokaryotes may have one or more reference or representative genomes per species.
-
- Reference genomes are selected based on assembly and annotation quality, existing experimental support, and recognition as a community standard (ex: Escherichia coli str. K-12 substr. MG1655) or of clinical importance (ex: Escherichia coli O157:H7 str. Sakai or Mycobacterium tuberculosis H37Rv).
- Representative genomes are assigned to type strain assemblies if there is no current reference genome or if one other than the reference is scientifically significant and exhibits strong sequence diversity as compared to the assigned reference genome(s) (ex: Mycobacterium avium subsp. paratuberculosis K-10 or Streptococcus thermophilus JIM 8232).
-
- Reference genomes are selected based on assembly and annotation quality, existing experimental support, and recognition as a community standard or of clinical importance (ex: Aspergillus fumigatus Af293).
- If there are no reference assemblies for a particular eukaryotic species, then RefSeq will select a Representative genome from the highest quality GenBank assembly (ex: Schistosoma mansoni ASM23792v2).
For more information: https://www.ncbi.nlm.nih.gov/assembly/help/
Key NCBI Resources for this Exercise
NCBI (Genome) Assembly database - a repository of genome sequence assemblies with information about submitters, statistics and links to the actual sequences.
NCBI Datasets has just recently added two new page types to display the above data!
Taxonomy pages - list information about the organism and related data at NCBI, as well as some summary information about a reference or representative genome.
Your Turn: Learn about the pathogen's genome!
Click below if you need a hint on what organism you found:
Identified viral isolate

Identified bacterial isolate

Identified fungal isolate
Find the Genome record for your pathogen
- Use the Search menu pull-down selector for Genome or go directly to the Genome database homepage (https://www.ncbi.nlm.nih.gov/genome) and begin typing in the name of the pathogen into the search text box.
As you type, you'll see "autocomplete" display some names to help you!
(This is based on information in our Taxonomy database - which we'll discuss in exercise 3.) - Click on the name of the pathogen you are looking for.
You may get directly to a record page for an organism or to a list of possible organisms. If you get the list, click on the name of the one you'd like to focus on.If you need it, you can click here to get to a link for the pathogen record page.
-
-
-
- About the organism
- A summary of information for the organism's genome
- Links to this organism's data in other NCBI databases
-
-
To begin, let's focus on the Genome section.
| NOTE: For those of you familiar with the old Genome & Assembly database records, they are still available for a time. You can access the old Genome page and link to related Assembly records with: View the legacy Genome page. |
| Download: You can quickly get metadata and various sequence datasets for this particular genomic assembly! |
Learn more about the reference or representative genomic assembly
If you need it, you can click here to get to pathogen genome page.
-
-
-
-
Summary
-
Assembly statistics
-
Assembly methods
-
Assembly details
-
Chromosomes - to access the sequence records, visualize or download a metadata table
-
Revision history - to see if/when the page has been updated
-
-
-
What can you learn about this particular genomic assembly?
Quickly download sequence datasets for your reference or representative genomic assembly
At the top of this Genome page is a very convenient Download button!
It gives you all sorts of options for what you can get for this particular genomic assembly.
What kind of data might you want to get?
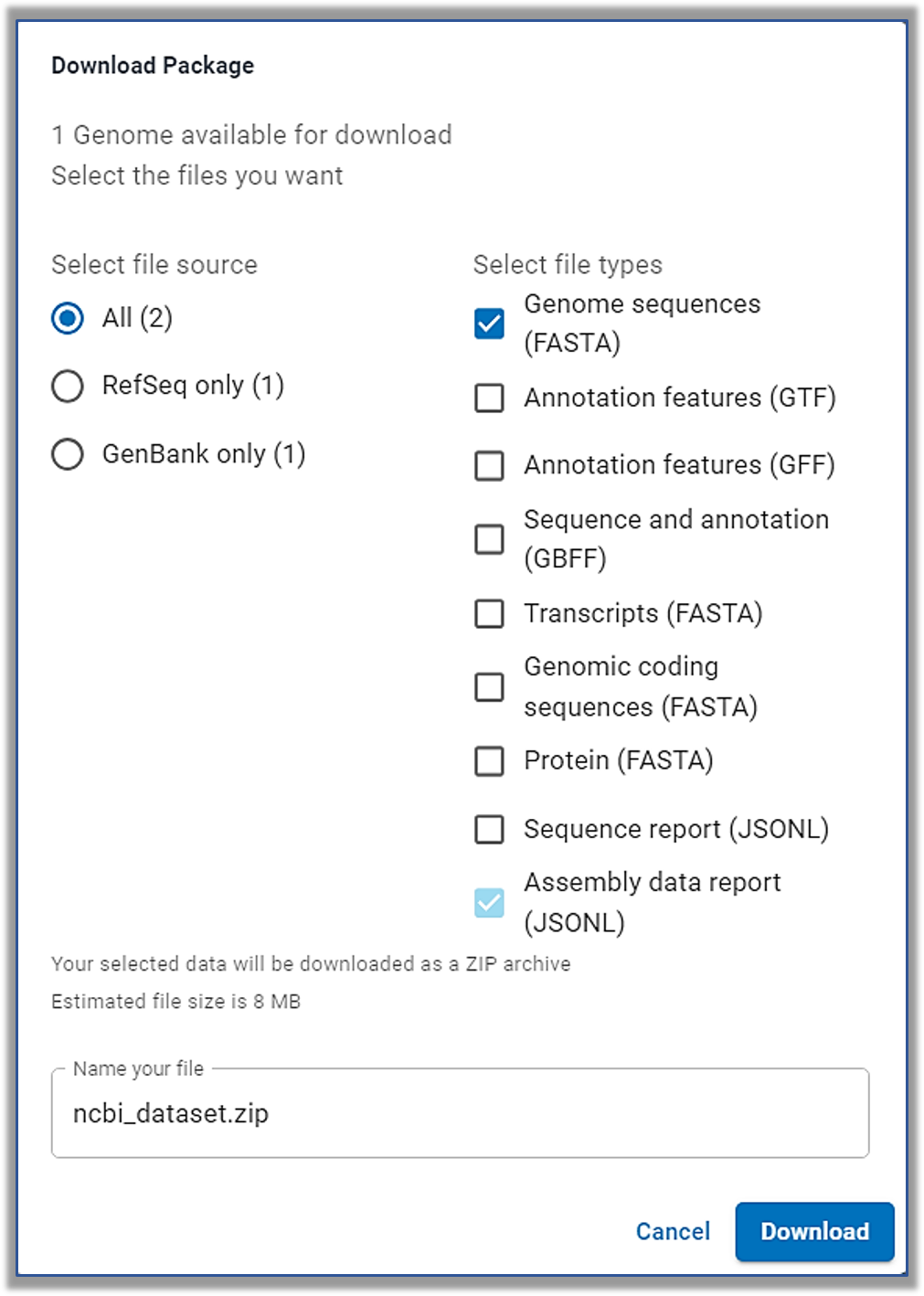 Learn more about the files. |
NCBI Datasets Genome Data Package Contents Dataset catalog: a list of each data file contained within or referenced by the package, along the content type and location for each.
Annotation Files in the following formats: Genome GBFF , Genome GFF3, or Genome GTF |
Explore genome annotations in a graphical viewer
-
-
Any nucleotide or protein sequence, including viral or bacterial genomic sequences or a eukaryotic nucleotide sequence, can be explored in the NCBI Sequence Viewer. This is available for any sequence record by clicking on "Graphics" or "Graphical View".
-
For eukaryotic genomes, we've expanded the types of data that can be accessed, displayed and compared with the NCBI Genome Data Viewer (a.k.a. GDV).
-
-
- On the NCBI Datasets Genome page, scroll down to the Chromosome section.
Here you can see each of the individual nucleotide chromosome records and their accessions as well as a little metadata for each chromosome.
- On the NCBI Datasets Genome page, scroll down to the Chromosome section.
| Note: You could also have clicked on an accession to go to the corresponding Nucleotide sequence record, and then clicked on "Graphics" to get to the same display! |
| As fungi are eukaryotes, you could also have clicked on Genome Data Viewer to use that particular browser. This enables you to search the entire genomic assembly and jump back between different chromosomes. Feel free to try this one on your own! |
Some things you can do with this genome browser:
-
-
- Use the slider to zoom in to see the annotations more clearly, or click an drag in the ruler bar to zoom into a region, or search with an annotated accession or name of a gene to zoom in directly to it....try this!
- for the viral pathogen: try searching with H (this encodes hemaglutinin - promotes viral infectivity by binding and aiding in cellular entry).
- for the bacterial pathogen:try searching with gyrA (this encodes DNA gyrase - regulates DNA replication and when mutated confers resistance to the antibiotic ciprofloxacin).
- for the fungal pathogen: try searching with CJI96_0001351 (this encodes FKS1 - a β-1-3-glucan synthase and when mutated confers resistance to the antifungal caspofungin)
- Use the slider to zoom in to see the annotations more clearly, or click an drag in the ruler bar to zoom into a region, or search with an annotated accession or name of a gene to zoom in directly to it....try this!
-
-
-
- Click the colorful button
 to quickly toggle display of the gene product (ex: protein) bars.
to quickly toggle display of the gene product (ex: protein) bars. - Place your cursor over a displayed bar and a pop-up a window with annotation and links to more information about that feature will appear.
- There’s a whole lot of documentation and several tutorials to help you learn how to use this viewer. To start, click on the “?” icon in the upper right-hand corner of the viewer.
- Click the colorful button
-
There are other workshops that help you learn to use this
and lots of video tutorials on YouTube!
see below
Take-away message!
-
- Search in the Genome for a Reference or Representative Genome assembly record.
-
- Simply click on the blue Download button!
-
- Viral and bacterial genomes can be Graphically viewed in the Sequence Viewer
- Eukaryotic chromosomes (such as fungal sequences) can be Graphically viewed either in the Sequence Viewer or the full assembly can be displayed in the Genome Data Viewer.
For more advanced work....
- NCBI YouTube Tutorial Playlist for Sequence Viewer
- NCBI YouTube Tutorial Playlist for Genome Data Viewer
- NCBI Virtual Workshop material for: "Using NCBI's Genome Data Viewer to Visualize Eukaryotic Genome Data" (May 26, 2022)"
- Prokaryotic Annotation Pipeline (PGAP) - this is available through a web interface, can be downloaded from GitHub or used in "the cloud"!
- Viral Annotation DefineR (VADR) - an application for viral annotations downlodable from GitHub or usable in "the cloud".
- NCBI Submission Portal - the best place to go to learn about and begin to submit data to NCBI!
- Entrez Programming Utilities (EUtils) - the NCBI-wide set of APIs for accessing and downloading NCBI data
- Entrez Direct (EDirect) - the NCBI-wide command-line tool for accessing and downloading NCBI data
- NCBI Datasets - a new, quick dataset resource with it's own command-line tool as well as programming utilities (APIs and Python- & R-related resources)
Last Reviewed: July 27, 2023


 Don't forget: You may need to search with [Candida] auris, not Candida auris. (
Don't forget: You may need to search with [Candida] auris, not Candida auris. (