Exercise 4: Find more information, including experimental studies with data, for research on the pathogen
It is often helpful for researchers to find more information about a pathogen for their work. However even knowing what is available can be a challenge. NCBI cross-references all molecular data based on the submitter's provided data source taxon which is then indexed and linked with records in the NCBI Taxonomy database. The number of records within each database for a taxon is quickly visible on a summary record - which enables researchers to click through and quickly retrieve those records for further examination.
In addition to sequence data, the Taxonomy records can include links to several different types of experimental study databases. Researchers are often interested in finding genotype/genetic variation data, gene expression studies or biological activity assays to include in their work on understanding their pathogen's biology as well as for developing diagnostics or therapeutics to assist the clinical community.
You will search and explore in NCBI's Taxonomy database to find information relevant to the pathogen. Then, you will identify where you can retrieve relevant experimental study data that may be helpful for further study of this organism.
Background
Taxonomy – central point of organization for NCBI’s molecular dataNCBI stores a vast amount and varied types of information for all sorts of organisms. The NCBI Taxonomy database was originally created to catalog and cross-reference GenBank sequences by their source organisms in a framework based on classical taxonomic levels (Kingdom, Phyllum, Class, Order, Family, Genus, Species) and at additional levels in between and below.
-
- Information in this database is manually curated in consultation with taxonomic experts, including:
- International Code of Virus Classification and Nomenclature
- International Code of Nomenclature of Prokaryotes
- International Code of Nomenclature for Algae, Fungi & Plants
- International Code of Zoological Nomenclature
- Information in this database is manually curated in consultation with taxonomic experts, including:

-
- Data is continually reviewed and updated.
Data in Taxonomy now includes non-traditional taxons such as communities of organisms, for example: microbiomes from environmental or host-organism isolates.
| PLEASE NOTE: Taxons included in NCBI Taxonomy are listed because someone submitted a nucleotide sequence to NCBI. This database does not contain all organisms known in our world. |
| NCBI is known for linking related data within and across databases. All molecular data is connected by data's source organism and NCBI Taxonomy provides the critical framework for this. Being able to find all related data at NCBI from a single organism page makes it easy to take the first step of really finding exactly what you are looking for, such as:
|
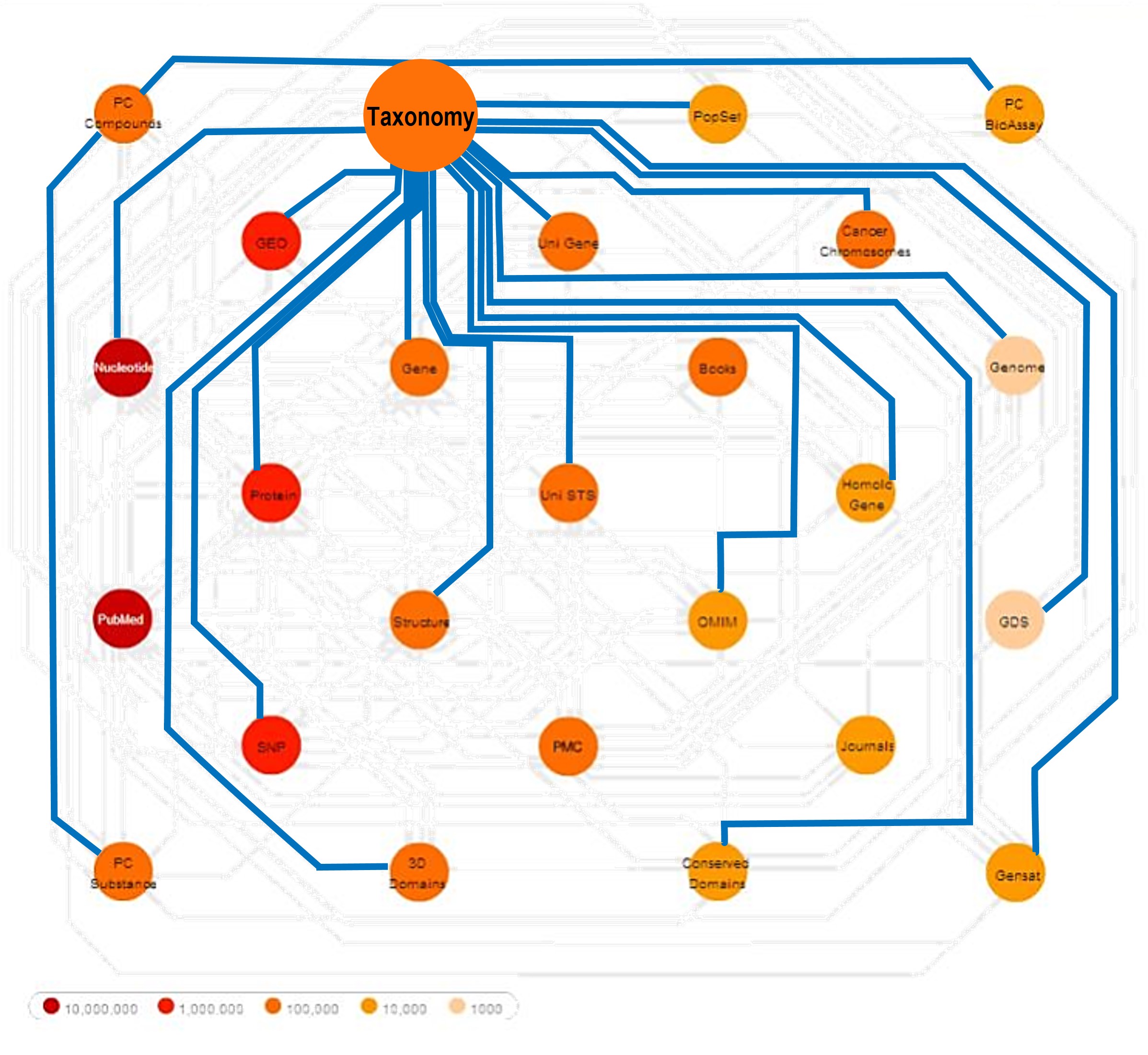 This is a very old (but pretty cool) image of how Taxonomy links related records across various NCBI databases. |
Key NCBI Resources for this Exercise
The NCBI Taxonomy database was created to provide structure for NCBI’s molecular data - beginning with GenBank. The taxon records display references for scientific naming rationale, common names, and provide links to additional taxonomy resources as well as relevant records across NCBI databases.
The NCBI Taxonomy Browser was developed to be able to view both the taxonomic hierarchy and types of available data in NCBI's databases simultaneously.
The NCBI Taxonomy Browser was developed to be able to view both the taxonomic hierarchy and types of available data in NCBI's databases simultaneously.
Your Turn: Find more relevant data for the pathogen for future studies!
Use the name of your patient's pathogen to begin your search following the steps below.
Click below if you need a hint on what organism you found:
Identified viral isolate

Identified bacterial isolate

Identified fungal isolate
Explore the Taxonomy database for information about the pathogen
-
- Search the Taxonomy database (https://www.ncbi.nlm.nih.gov/taxonomy/) with your pathogen name. Then, click a name to see a hierarchical list of all organisms underneath this taxonomic node in the Taxonomy browser tool.
If you need it, you can click here to get to a link for the pathogen Taxonomy browser view.
What do you see?
| If you are interested in communities of organisms with "metagenomes": NCBI also has received and provides sequence data from environmental isolates or “metagenomes”. NCBI metagenomes are a defined, pseudo-organismal eco-system consisting of the genomes and genome products of many individual organisms, such as a microbiome. You can search the Taxonomy database with the term "metagenomes" to see the types of community isolates that people have submitted to NCBI sequence databases. |
2. There is a control panel at the top of this view which enables you to adjust what you are looking at.
-
-
- You can set the start level for the diplayed hierarchy by clicking on names within the flat taxonomic hierarchy shown just above the displayed hierarchy. Note: “Mouse over” the name to show you the classification name for that level.
- In the top of the control panel you can adjust the the number of hierarchy levels shown by changing the number next to "Display" and then clicking the Display button.
- Perhaps more importantly, you are able to display the number of relevant datasets from various NCBI databases or projects directly next to the names listed in the hierarcy.
- Check boxes next to database names you are interested in and then click the "Display" button to reveal color-coded number of records next to each corresponding organism shown within the structured view!
-
For example, at the top of the page, check the blue Nucleotide, red Protein, pink Genome, purple Gene and/or green Structure, then click the "Display" button.
For your particular pathogen of interest, how many records are available in each database?
Learn more about the pathogen, including types and amounts of related data at NCBI
-
- Select a particular taxon that you are particularly interested in, by clicking on the name to see the Taxonomy summary page. Note: You may need to scroll or use the web-browser’s “Find in page” function (Ctrl+F) to search for it in the list.
If you need it, you can click here to get to a link for the pathogen Taxonomy record page.
2. Explore the Taxonomy summary page to see all of the information available for the pathogen. Every organism has it's own collection of information, some with more - some with less....
| NCBI’s Taxonomy group works with outside experts and our Reference Sequence “Genome Champions” to curate these records. Notes with information or references about the official designations are often added in addition to common names, references and helpful links to eternal resources. We're always looking to add information that researchers would find helpful. Is there any additional information that you would like to see? Let us know! |
3. Look on the right-side of the page for the data table listing the number of records and provides links to databases for which we have records for this particular organism. These numbers should correspond with those shown within the hierarchy view.
Some of these may be databases you recognize, some - maybe not!
While you can certainly initiate a search in any of these databases directly, often the easiest way to find records that are specific for your organism is to start from this Taxonomy page!
Find an interesting experimental study for the pathogen
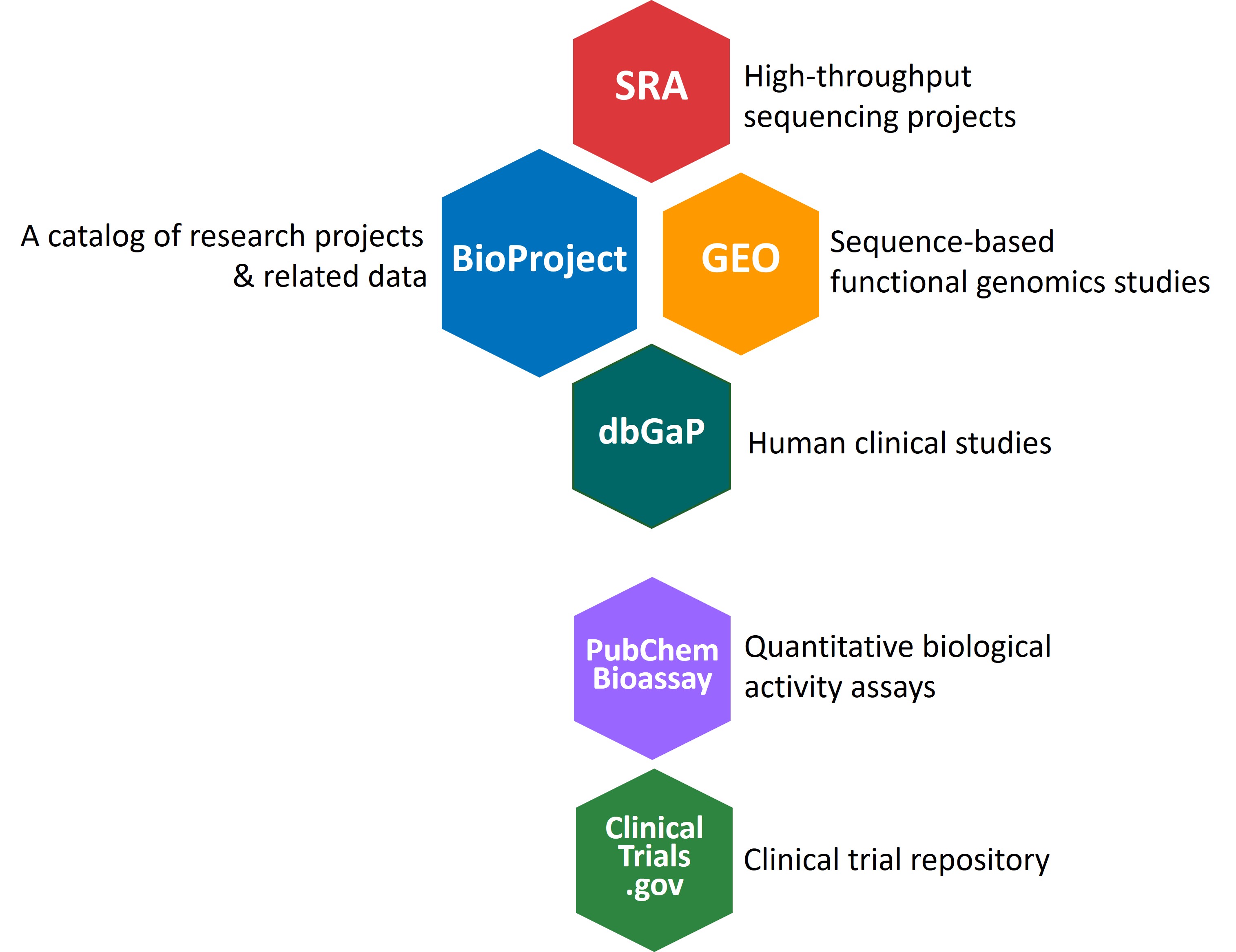
A quick overview of NCBI experimental study databases that might be of interest to you:| BioProject - a catalog of sequence-based research projects & related data Sequence read Archive (SRA) - Studies including genome resequencing for assembly & annotation (DNASeq, RNAseq, ChIPSeq, etc.)
Gene Expression Omnibus (GEO) - Functional genomics studies including Genotype (Microarray/DNASeq), Expression (Microarray/RNASeq), and Epigenomics experiments. Database of Genotypes and Phenotypes (dbGaP) - Human Clinical Studies information and related data Other study databases not listed in BioProject: PubChem BioAssay - Biological activity assays (experiments measuring the impact of an exposure to a particular chemical or RNAi on cell growth, enzyme activity, receptor binding, etc.)
ClinicalTrials.gov - Human clinical trials (a listing repository with result information included for some trials) |
|
What type of experimental study data would you like to search for?
Take-away message!
If you are looking for what taxonomic information or biological data is available based on a specific organism or taxon...
-
- Start by searching with the organism's name in the NCBI Taxonomy database!
To explore what data is available for your organism and others closely related to it...
-
- Use the Taxonomy Browser to see the number of available records for your choice of databases for each organism in the context of the hierarchical view.
To quickly retrieve records for your organism or taxon in any NCBI biological database, for example - an experimental study database...
-
- Start with a Taxonomy Record and click to move to that database with records already retrieved specifically for that organism or taxon.
For more advanced work....
-
- Getting a bit of experience starting on the Amazon Web Services (AWS) Cloud with viral sequence data, an NCBI Workshop "An Introduction to NCBI Cloud Computing for Virologists" (February 8, 2022)
- A NCBI Workshop showing how to mine pathogenic microbe data in the Pathogen Detection Project resources: "Identifying Clinically Relevant Genes in Bacterial Genomes" (May 19, 2022)
- For those of you interested in identifying members of communities with metagenomes: an NCBI Workshop "An NCBI Guide to Finding & Analyzing Metagenomic Data" (October 28, 2021)
-
- We are working on creating more advanced workshops for accessing and doing preliminary work with NCBI experimental study data, such as "Analyzing & Visualizing NextGen Reads with Magic-BLAST" (December 13, 2022)
Please stay tuned to the NCBI Outreach Events page for announcements of new workshops!
Last Reviewed: July 27, 2023


 Don't forget: You may need to search with [Candida] auris, not Candida auris. (
Don't forget: You may need to search with [Candida] auris, not Candida auris. (