Appendix
Homology: Orthologs and Paralogs
Homology refers to biological features including genes and their products that are descended from a feature present in a common ancestor. Homologous features such as genes are referred to as homologs (or homologues if you follow British spelling).
Homologous genes become separated in evolution in two different ways: separation of two populations with the ancestral gene into two species or gene duplication of the ancestral gene within a lineage.
- Genes separated by speciation are called orthologs.
- Genes separated by gene duplication events are called paralogs.
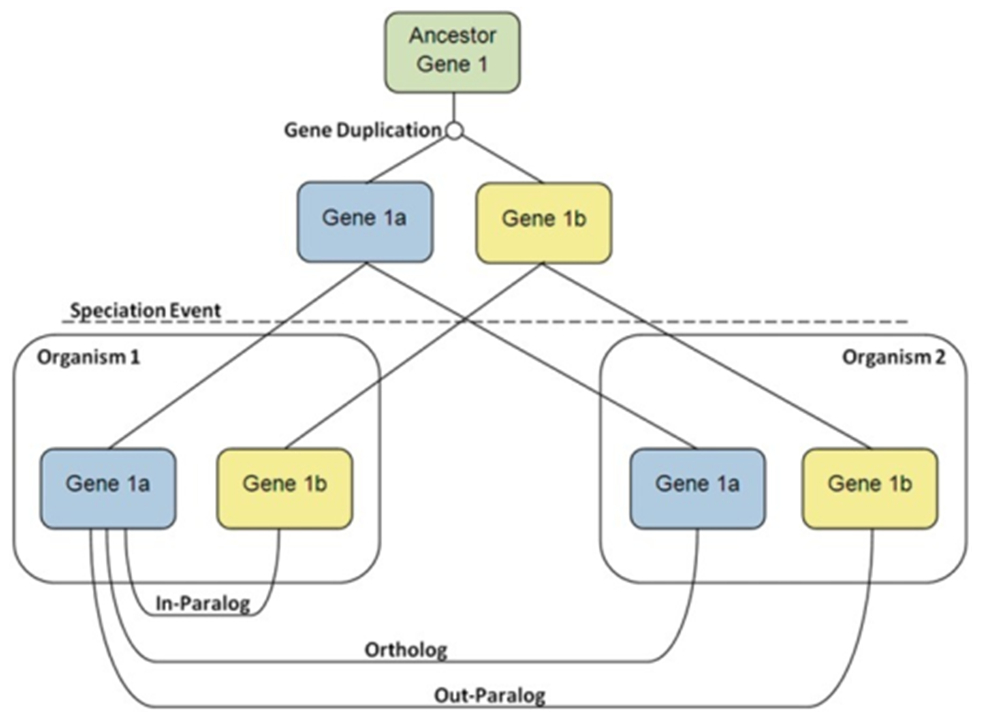
The process is shown in the diagram below.
Gene 1 in the ancestral species undergoes a duplication event generating Gene 1a and Gene 1b. The ancestral species splits into two species, each with its own copy of Gene 1a and Gene 1b
- Gene 1a in species one is the ortholog of Gene 1a in species two.
- Gene 1a and Gene 1b are paralogs.
- All four genes are homologs.
How BLAST works (over simplified)
- BLAST finds short seed matches (word hits) between a query sequence and a database sequence.
- The word size parameter controls the size of these initial matches and affects speed and sensitivity. The default nucleotide search program, megablast, uses larger word sizes than the traditional blastn program, which is partly why megablast is faster than blastn, but less sensitive
- Intial matches are extended as alignments until the alignment score declines below a certain threshold.
- BLAST then attempts to extend these ungapped alignments by including gaps.
- The gap open and extend penalities affect the size, number of gaps and the lengh of the alignments
- BLAST ranks and filters matches by how unlikely they are, returning — those that wouldn't be expected by chance
- The expect value parameter sets the stringency of this filtering
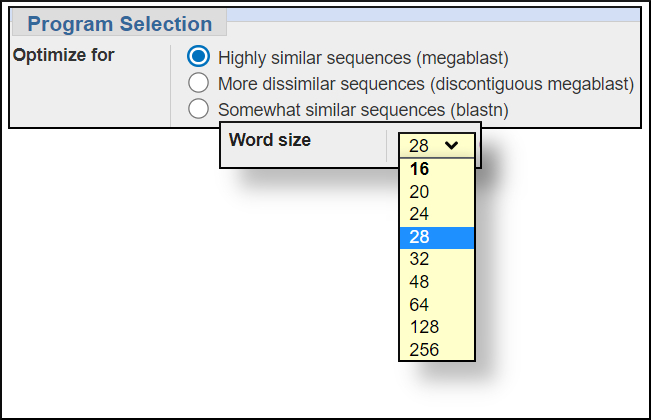
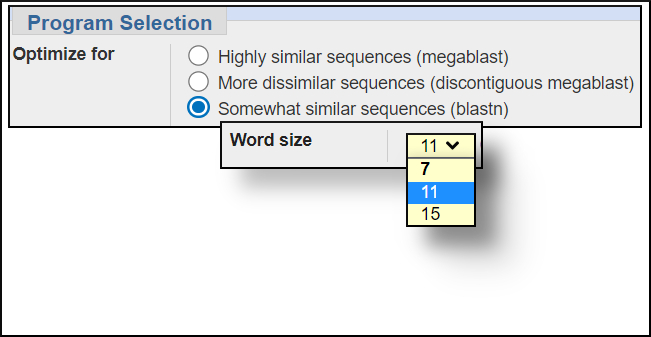
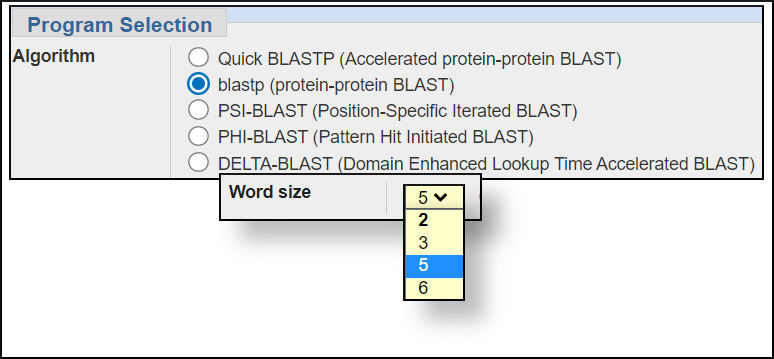
Word size settings
You can change the word size under the Algorithm parameters of the BLAST submission form. In general, larger word sizes increase the speed of a BLAST search and decrease the sensitivity, smaller word sizes make the search run slower but increase the sensitivity.
The available settings for the BLAST programs are shown in the images below. In this workshop we won't change any of the default settings but will see the difference in the results for the same search when using megablast and blastn.megablast word size options
blastn word size options
blastp wordsize options
BLAST Scoring and Statistics
Position-independent Scoring
Traditional BLAST uses position-independent scoring: the same substitution gets the same score all any position in the alignment.
Nucleotide Scoring
Nucleotide alignments use an identity scoring system, a simple match mismatch scoring system with a positive score for match and a negative score for a mismatch and gap open and extend penalties. The image below shows how BLAST scores and represents a nucleotide alignment.
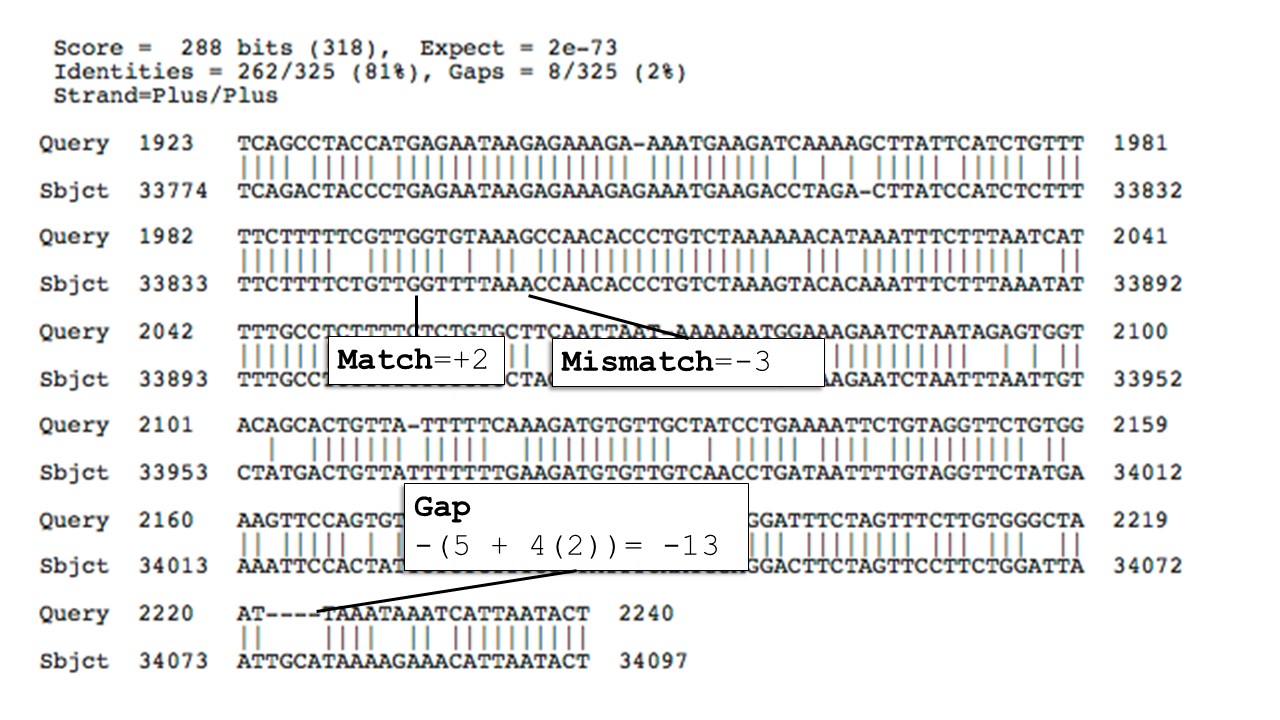
You can use BLAST 2 Sequences to see a megablast alignment between a human insulin transcript (NM_000207.3) and a predicted insulin transcript (XM_043971863.1) from the colocolo opossum.
- Set up the search and run it by clicking the BLAST button.
The above alignment was produced by the megablast program, which is less sensitive (but faster) than blastn.
- Set up the blastn search and run it.
Do you differences in the alignment and score using the more sensitive program?
Protein Scoring
Protein alignments use a scoring system based on frequencies of amino acid substitutions in related proteins. The default scoring matrix is BLOSUM62, shown below. The BLOSUM series uses observed substitution frequencies in ungapped alignment blocks of related proteins. BLOSUM62 includes information up to 62% identity. Experiment have shown that this is the best general scoring system. Other available matrices for protein BLAST include several from the BLOSUM series tuned to different distances and several from the PAM series.
The numbers in BLOSUM62 are log odds ratios of the observed substitution frequency to the background frequency. Substitutions that occur more often than expected by chance have positive scores, those that occur less often than chance have negative scores, and those that occur at the background frequencies get a score of zero
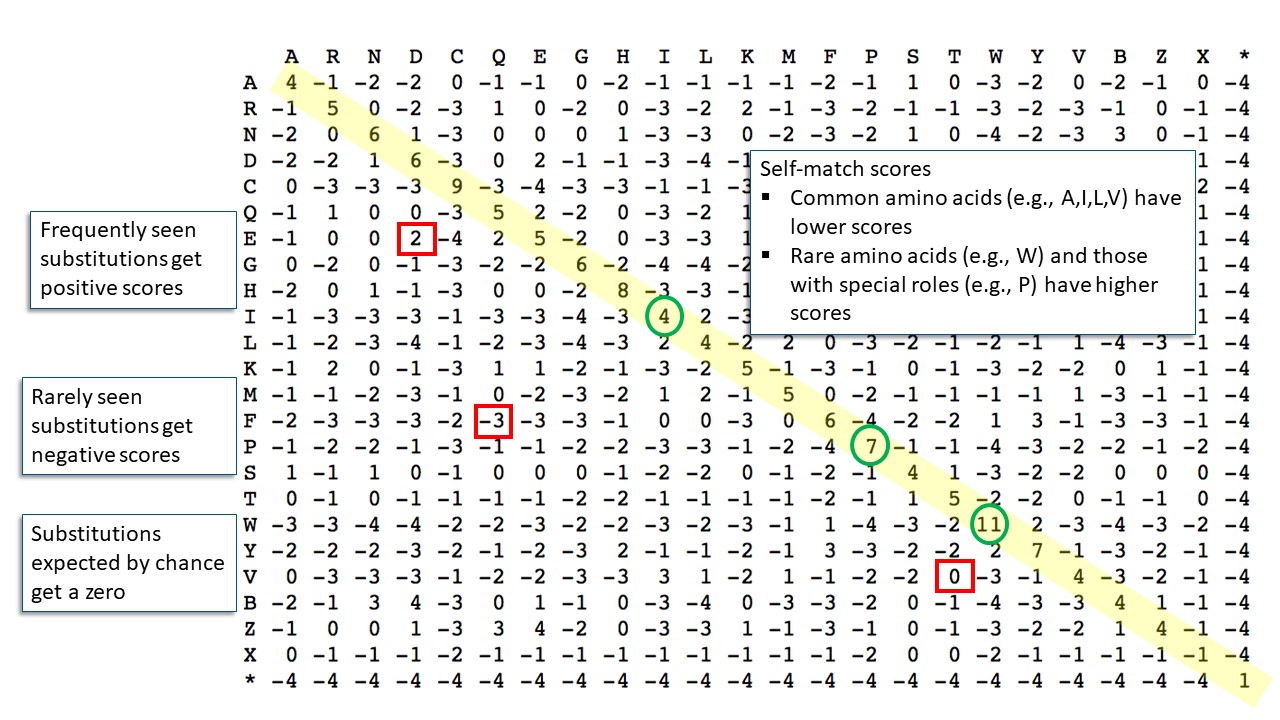
It's easy to understand the BLOSUM62 scores based amino acid chemistry and protein structure. Amino acid substitutions with side chains of similar size and chemistry have positive scores (e.g., aspartate (D)/glutamate (E)). Those involving dissimilar side chains have negative scores (e.g., phenylalanine (F)/glutamine (Q)). Self-substitution scores are along the diagonal and, in part, reflect the abundance of the amino acids. Rare amino acids such as tryptophan (W) have relatively high scores. Common amino acids such as valine (V), leucine (L), and isoleucine (I) have lower scores. The relatively high self-scores for proline (P) and glycine (G) may be because these amino acids often have special roles in determining protein structure. Keep in mind though that the substitution scores in the BLOSUM matrices are based on observed frequencies not on any predictions from amino acid properties.
The image below shows how BLAST scores and represents a protein alignment.
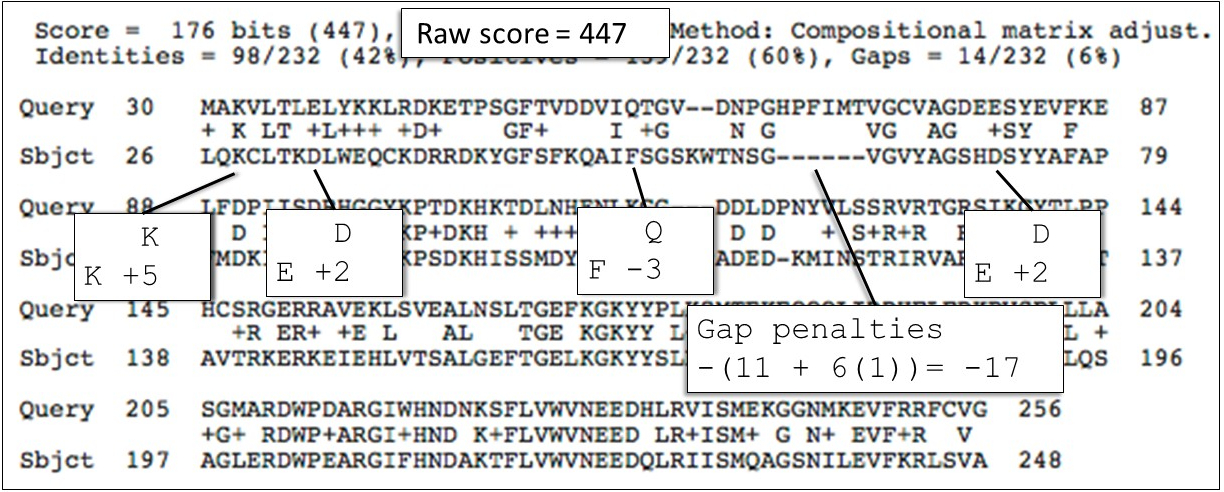
You can use BLAST 2 Sequences to see a blastp alignment between the human creatine kinase M protein sequence (NP_001815.2) and a bacterial arginine kinase protein (MCP4285491.1).
- Set up the search and run it by clicking the BLAST button.
Position-dependent scoring
The position independent scoring systems make the unrealistic assumption that every position in a protein or nucleotide sequence is equally likely to change. Position specific scoring strategies described next do a better job of modeling real biological sequences and increase sensitivity
Specialized BLAST protein programs such as PSI-BLAST and the Conserved Domain Database (CDD) Search (RPS-BLAST) generate or search a database of Position-Specific Scoring Matrices (PSSMs). In a PSSM the score for a particular substitution depends on the position in the alignment. This is a better model of proteins since it can represent the fact that amino acids that are directly involved in catalysis, substrate, cofactor, or partner interaction as well as those required for critical structural elements are less likely to change than others. PSSMs are generated from multiple sequence alignments either generated on-the-fly from a BLAST search in the case of PSI-BLAST or as a curated database of conserved domains used by CDD search. PSSMs are better at detecting more distant protein relationships than ordinary BLAST and can have a more direct relationship to protein structure and function.
You've used PSI-BLAST for one example in this workshop. CDD search runs by default in all of our protein examples and will show any conserved domains in your protein queries.
BLAST Statistics: The Expect Value
The sequence alignment scores we have been discussing imply a particular model of biological sequences. Based on that that model we can calculate or simulate the distribution of random (chance) alignment scores. So BLAST can provide information about whether an alignment score is likely to have come from that distribution of random alignment scores. The relevant statistic is called the Expect Value or e-value.
Expect value — for a particular match, the number of chance alignments expected with the same score or a better one.
The Expect value is an exponentially decreasing function of the score and is directly proportional to the search space. If the expect value is very much less than one, then the alignment score is not due to chance. However, if the expect value is near one or a greater number, it means the score may be due to chance, but doesn't mean it is.
Important: Since the Expect value increases with the database size, you should always search the smallest database that contains the sequences of interest. On the web, you can choose a smaller database from the menu and restrict the database using an organism limit or one of the exclude filters.

The Expect Threshold and Max Target Sequences
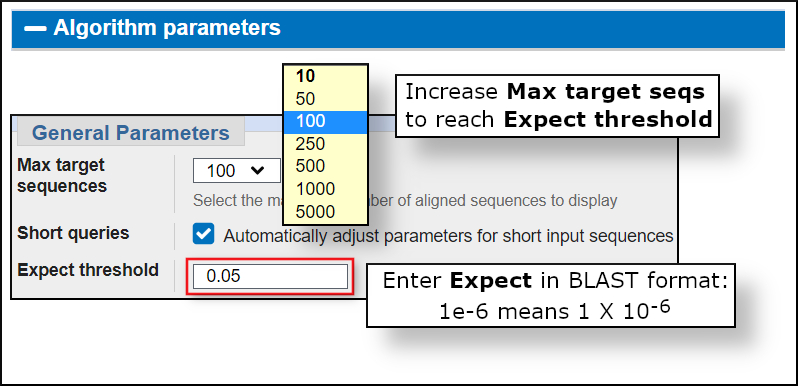
When running a BLAST search, you probably want to see all the significant matches. In other words, you want to make sure that you reach the expect value cutoff set for the search. The default cutoff (Expect threshold) is set to 0.05 for protein and nucleotide searches. However, the default number of matches returned is only 100. The number of matches is controlled by the Max target sequences setting. In most searches, you will need to increase this setting to see all significant matches. The Expect threshold and Max target sequences settings are under the expandable Algorithm parameters section of the BLAST form as shown in image below.
BLAST Databases
NCBI has a large number of databases available on the web service. You'll use only a few of them today. You select databases using the pull-down lists and radio buttons in the 'Choose Search Set' section of the BLAST submission form. Both the nucleotide page and the protein page have standard and experimental databases.
The nucleotide and protein pages have default databases called nr/nt and nr respectively. Other databases include useful subsets of these as well as separate databases with different content. Today you'll search the default databases as well as the NCBI RefSeq subsets. You'll also search a whale genome assembly that's available through a separate Genome BLAST page
Note: The sequence entries in the BLAST databases are non-redundant. (This is what the database name nr stands for.) Non-redundant means that identical sequences are represented by a single entry in the database. In the case of protein sequences, sometimes hundreds of sequences may be collapsed into a single entry. BLAST provides a way to access all of the identical proteins for a non-redundant sequence in the BLAST database.
Nucleotide
Standard Nucleotide Databases
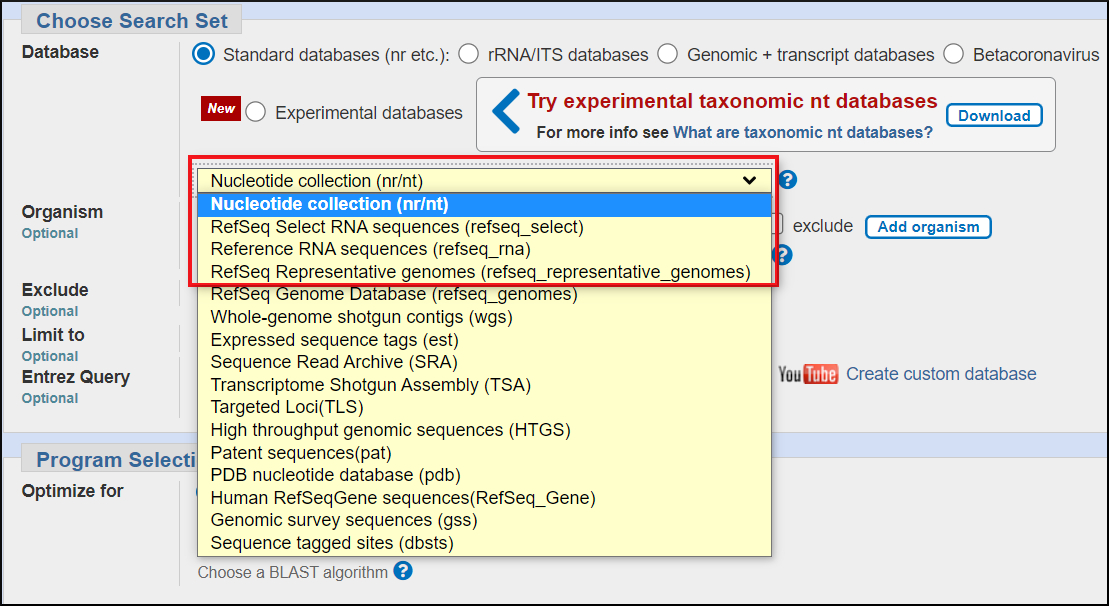
- The default database (nr/nt) contains traditional GenBank and RefSeq RNA sequences.
- less comprehensive than protein nr
- no RefSeq genome sequences and no eukaryotic genome assemblies
- no high throughput sequences (whole genome shotgun, next-gen reads from the Sequence Read Archive.
- Subsets of nr/nt
- RefSeq RNA
- predicted transcripts from the eukaryotic genome annotation pipeline
- high quality NCBI-curated transcript sequences
- RefSeq Select
- selected curated transcripts for mouse and human genes
- one transcript per gene.
- RefSeq Representative Genomes
- Highest quality NCBI curated RefSeq genome assemblies for a taxon.
- Example: the human GRC38 reference genome assembly.
- RefSeq RNA
Experimental Nucleotide Databases: Organism-based nt databases
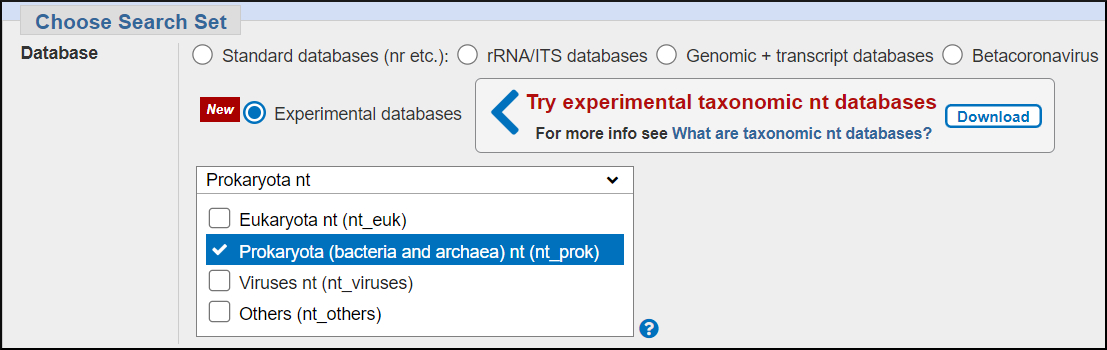
The very large — 96 million sequences and 1.3 trillion total bases, and rapidly increasing size of the nr/nt database makes it difficult to manage and maintain for both NCBI and outside people who use BLAST locally.
We're experimenting with ways to divide nt/nt into more manageable subsets. One strategy is to divide it by organism (taxonomy) into four databases.
- Eukaryota nt (77 million sequences and 836 billion bases)
- Prokaryota nt (8.4 million sequences and 207 billion bases)
- Viruses nt (10 million sequences and 217 billion bases)
- Others nt (919 thousand sequences and 1.9 billion bases)
Others nt includes metagenomic and artificial sequences.
These databases are on the web and available to download. We are interested in feedback on how useful they are.
Protein
Standard Protein Databases
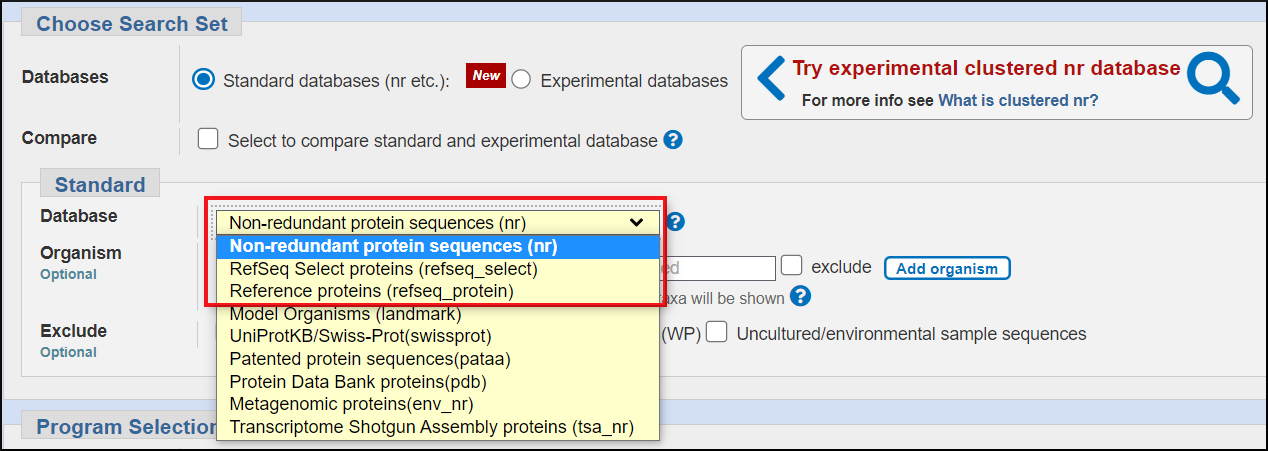
- The default protein database nr contains nearly all protein sequences available at NCBI.
- no patent sequences
- no wgs metagenomes and no transcriptome shotgun assembly proteins
- includes protiens from outside protein-only sources that are alsoavailable as separate databases.
- Subsets of nr
- RefSeq Protein
- coding region translations from RefSeq transcripts and RefSeq genome annotations
- RefSeq Select Protein
- coding region translation from mouse and human RefSeq Select transcripts plus coding regions from RefSeq reference and representative prokaryotic genome
- RefSeq Protein
Experimental Protein Databases: ClusteredNR
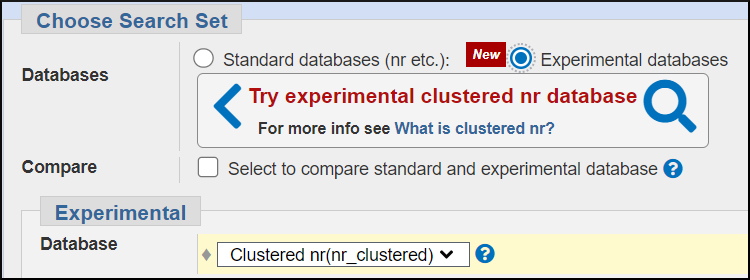
The protein nr database is also large — 595 million sequences and 234 billion residues, and growing rapidly. In addition, certain kinds of proteins and groups of organisms such as mammals and their associated bacteria are over represented and will dominate protein searches. This makes it difficult to identify more distant homologs in other species.
One strategy to collapse some of the over-representation is to form clusters at less than 100% identity.
The experimental ClusteredNR has clusters where the members are 90% identical and within 90% sequence length.This provides access to more distant matches and reduces the computational burden of searching entire nr since BLAST only searches the representative sequence for each cluster. ClusteredNR currently has 286 million sequences and 94 bilion total residues — about 40% the size of the full nr. You'll use this database to expand the results for a blastx search and as a way of incorporating more distant matches in a PSI-BLAST search.
Last Reviewed: August 1, 2023

