Protein BLAST, COBALT, and Protein Trees
We'll use the human muscle creatine kinase protein (NP_001815.2) to find homologous proteins in the RefSeq protein database. We can then make a multiple alignment and construct a protein tree. Because of the presence of multiple related proteins (paralogs) in the various species, the resulting tree shows both the phylogeny of the species and the phylogeny of the different genes and their products.
Finding human creatine kinase paralogs with blastp
Goal
Identify all creating kinase protein homologs in human using the RefSeq Select muscle creatine kinase protein as a query.
Search setup
Query sequence
Retrieve NP_001815.2 from the protein database and use the 'Run BLAST' link on the upper right to send the sequence to the BLAST form. Or go to the BLAST homepage and select Protein BLAST search and enter the accession number.
Search type
Keep the default blastp search.
Database, limits, and filters
Select the RefSeq Select proteins (refseq_select database). This database contains coding region translations for selected RefSeq Select transcripts for human, mouse, and rat — one per gene — as well as RefSeq prokaryotic genome protein annotations.
Use human (taxid:9606) as an organism limit.
BLAST program
Keep the default blastp program
Run BLAST!
Results
Descriptions
As with nucleotide searches the results are sorted by significance, Expect value. There are four distinct kinds of proteins here (U, M, S, and B). These are the products of different genes (paralogs) in human. There are five sequences though because there are two separate genes on chromosome 15 that produce the U-type proteins. We expect to see these same gene products in other vertebrates
Notice that this is the classic use of BLAST, identifying homologous proteins. In this case, they are homologs in the same species.
Graphic summary
As with the graphic summary in the nucleotide searches, the display shows how the database matches align to the query sequence. Notice that the U and S proteins don't align at the N-terminal because of their signal peptide sequences.
The protein Graphic summary also shows the results for a Conserved Domain Database search that searches a database of position specific score matrices and identifies conserved domains in the query sequence. It identifies the creatine kinase like domain and the phosphagen kinase superfamily as present in the query. Had this been an unknown protein sequence, you could have immediately assigned a probable function to it based on this result
Interpretation and conclusion
The RefSeq Select database is useful for quickly identifying homologs in human, rat, and mouse. In this case, we used it to identify homologs (paralogs) in the same species. RefSeq Select is useful in many cases because it remove the complications due to multiple isoform products.
Finding creatine kinase homologs in non-model RefSeq proteins, generating a multiple sequence alignment and a protein tree
Goal
Find creatine kinase protein homologs in non-model reference sequence and construct a protein phylogenetic tree from the results
Search setup
Use the 'Edit and resubmit' link to return to the search from. Use the same Query sequence and Search type.
Database, limits, and filters
Choose the Reference protein (refseq_protein) database. This database contains coding region translations of NCBI Reference Sequence mRNAs as well as Reference Sequence protein genome annotations for prokaryotic genomes
Remove the organism limit and use both the exclude XP_ and WP_ options to exclude gene models and the RefSeq prokaryotic proteins.
These settings restrict the RefSeq proteins to non-model RefSeqs that are based on submitted transcript sequences that have good experimental support. This will create a small database of protein products to construct a protein tree.
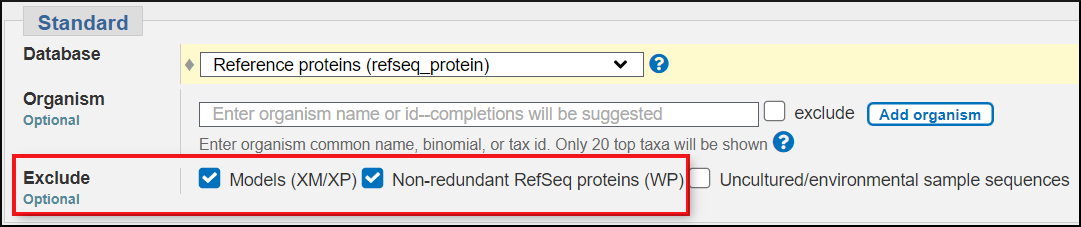
Run BLAST!
Results
Descriptions
We have 98 database sequence matches for a number of proteins and organisms. There are several different creatine kinase proteins and the related arginine kinases from invertebrate. There are no matches to prokaryotes because we excluded those from the search database otherwise there would be many matches to prokaryotic arginine kinases.
The Descriptions are sorted by Expect value. The largest (least significant) is 3e-13 for an uncharacterized Drosophila protein. But all are significant matches
Taxonomy
The Taxonomy tab shows wide range of animals including mammals, bony fishes, cartilaginous fishes, other vertebrates, as well as tunicates, a sea urchin, insects, and a nematode.
Using COBALT to make a multiple sequence alignment and a phylogenetic (protein) tree
Go back to the Descriptions
You can make a distance tree right from the BLAST results using the link at the top of the Descriptions section. But to get a more accurate alignment follow the 'Multiple Alignment' link. This will run a full-length multiple sequence alignment using COBALT. Unlike BLAST, the COBALT tool produces a full multiple sequence alignment (MSA) that includes all positions of all sequences. As with all multiple sequence alignment programs, you need to provide the tool with sequences that are related. Here you used BLAST to provide COBALT with a set of related sequences.
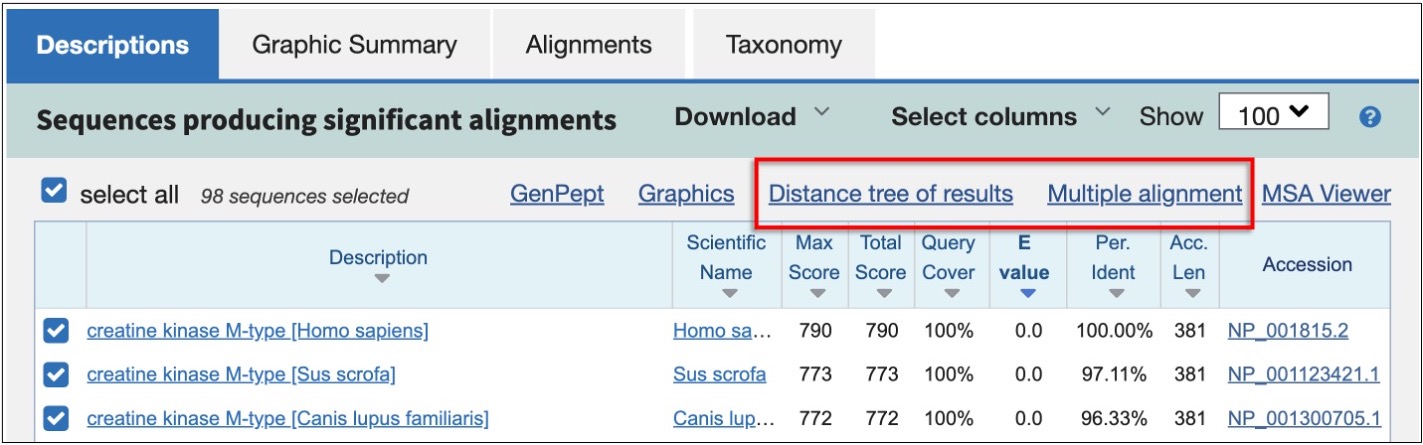
Inspect the alignment using the viewer at the top of the results or the text version at the bottom.
You may want to unselect the following six aberrant sequences and re-align before proceeding with making the phylogenetic tree:
-
- NP_999687.1 creatine kinase, flagellar [Strongylocentrotus purpuratus]
- NP_001107070.1 zgc:172076 [Danio rerio]
- NP_729446.1 arginine kinase 1, isoform A [Drosophila melanogaster]
- NP_650501.1 uncharacterized protein Dmel_CG4546 [Drosophila melanogaster]
- NP_726252.1 uncharacterized protein Dmel_CG30274 [Drosophila melanogaster]
You can download the MSA in various formats from the top of the page if you want to edit and analyze it locally.
Generate the phylogenetic tree from the MSA through the link at the top of the COBALT results.
The Tree Viewer shows the protein tree built from the MSA. To expand the viewer, click the plus sign [+] in the upper left hand corner of the viewer. The arginine kinases and creatine kinases are in their own distinct branches. The collapsed branch with the yellow highlight contains the query sequence
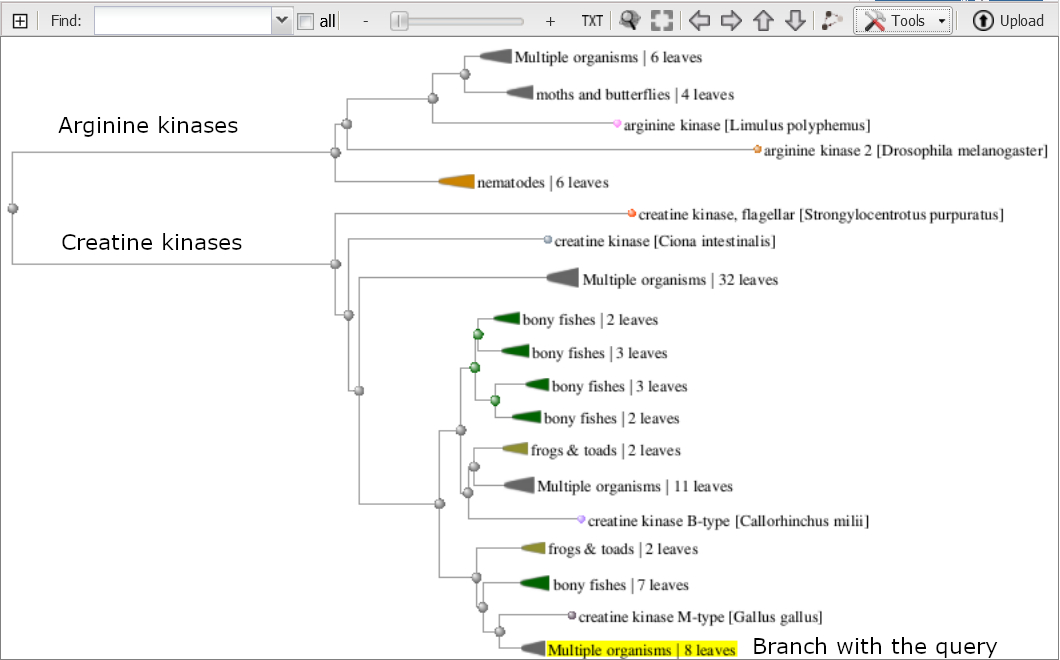
Use the 'Tools' menu to 'Expand all' branches and navigate to the creatine kinase section of the tree. Notice that the different creatine kinase genes each form their own distinct group (M,U,S,B). Within each group the branching pattern broadly reflects the phylogeny of the organism groups in each as shown below for the M-type creatine kinases.
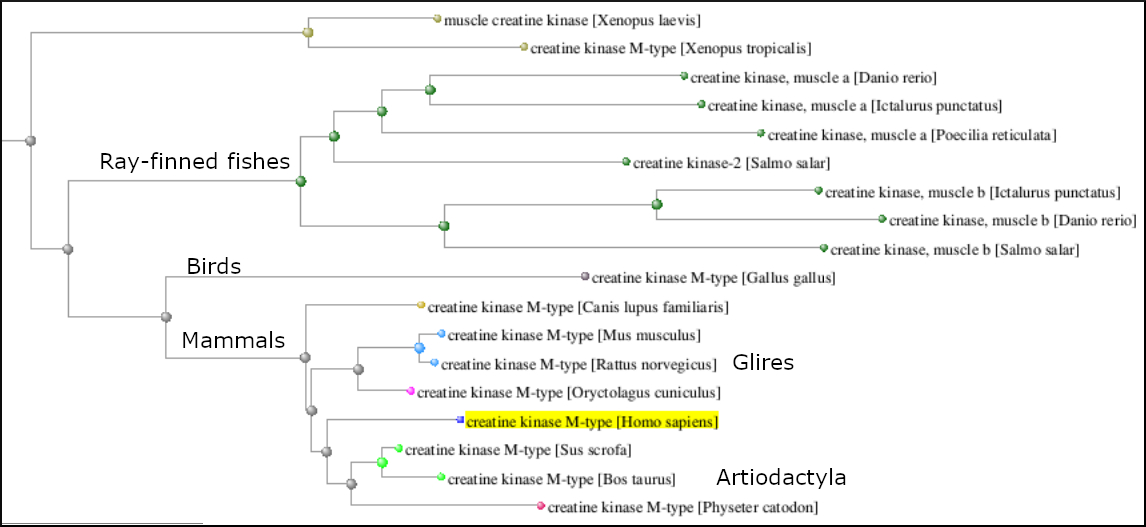
In this example you used a limited set of sequences, NP_ RefSeqs. You can work with a more complete set of data if you run this search without the XP_ filter. You would need to limit to a smaller taxonomic set, such as a group of mammals (Laurasiatheria, Euarchontoglires, etc.)
As with the MSA, you can download the text tree file in different formats to analyze it locally or generate tree graphics.
Interpretation and conclusion
Using a limited database, you collected a set of creatine kinase homologs from across the animal kingdom. The tree shows both the branching pattern caused by phylogeny and the branching pattern from gene duplications
Saved non-model RefSeq results
Last Reviewed: July 5, 2023

