Nucleotide Search and Database Examples
The creatine kinase gene family
Creatine kinases are a small family of related genes whose protein products generate phosphocreatine, which serves as a source of high energy phosphate for rapid regeneration of ATP. This process is important in cells with rapid ATP turnover such as muscle cells. Humans have five distinct creatine kinase genes that are expressed in various cell types. Some are targeted to the cytosol and some to the mitochondria.
You'll use the creatine kinase genes and their products in several examples today to learn how to use BLAST to find homologs and explore the relationships among them.
Finding creatine kinase matches in the default nucleotide database (nr/nt)
Goal
Find CKM transcript homolog matches in nr/nt to a specific taxonomic group (whales & dolphins).
Search setup
Query sequence
Use the human muscle creatine kinase RefSeq Select transcript, NM_001824.5.
Search type
Use nucleotide (blastn) search.
Database, limits, and filters
Use the default database nr/nt with no modifications
The default database (nr/nt) contains traditional GenBank and RefSeq RNA sequences and is defined more by what isn't included than what is included.
-
- no RefSeq genome sequences and no eukaryotic genome assemblies
- no high throughput sequences (whole genome shotgun, next-gen reads from the Sequence Read Archive, etc.).
nr/nt is a very large and poorly defined database making it difficult to find all matches in manageable set of results.
BLAST program
Use the default BLAST program (megablast).
Algorithm parameters
Increase the Max target sequences to 500.
Run it!
Results
Descriptions
There are over 500 hits to a wide variety of sequences (genomic and transcript) from a large number of different organism. All matches have an e-value of zero. Even with 500 target sequences you probably haven't reached the Expect threshold setting and are missing significant hits.
Graphic Summary
There are a wide range of query coverages. Notice that some matches show a dark line in the graphic. The dark lines represent regions of the the subject sequence that don't align to the query. If you mouse-over these notice that some are genomic sequences (e.g., Homo sapiens isolate CHM13 chromosome 19, CP068259.2) and the dark lines represent introns in the subject
Alignments
The first several matches are to identical and nearly identical sequences from Primates. Jump to CP068259.2 in the Descriptions to see that the multiple matches to the chromosome 19 exons are shown together headed by the highest scoring match.
Taxonomy
Except for the synthetic constructs, everything in this output is from a vertebrate. There are many other significant matches that are missing.
You can use the Filter results box at the top to find results for a specific taxon. Filter for whales & dolphins (cetacea). You have 15 matches to cetacean sequences. All are NCBI Reference Sequence transcript sequences. All are also muscle creatine kinase orthologs. None of the products from paralogs are in the output.
Interpretation and conclusion
The large default database limits our ability to find all matches. You can re-run the search in the next example using a smaller database to find a more complete set of results.
Saved ResultsUse a smaller more focused database to find additional matches for human muscle creatine kinase
Goal
Find a complete set of megablast matches in whales for the human muscle creatine kinase transcript. In the last example you weren't able to see all matches because of the large number of results from nr/nt . You can use a smaller database and an organism limit to get a better set of matches.
Search set up
Use the same Query sequence and Search type as in the first example. You can use the 'Edit Search' feature on the BLAST results to return to the submission form with the query sequence loaded.
Database, Limits, and Filters
Chose the Reference RNA Sequences (refseq_rna) database.
This database, which is a subset of nr/nt contains:
-
- predicted transcripts from the eukaryotic genome annotation pipeline
- high quality NCBI-curated transcript sequences
Use the Organism box to restrict to whales and dolphins (taxid:9721). This organism limit on the search pages reduces the database size before the search to focus on the organisms of interest.
Run it!
Results
The Descriptions now shows 24 database sequence matches including transcripts from the creatine kinase B paralog. Since the maximum target sequences was set to 500 and BLAST only returned 24, we know that these are all the hits that BLAST can find under these conditions (megablast, Reference RNA sequence database, whales and dolphins organism limit). We'd need to use a more sensitive blastn or protein level search to find the additional paralogs that are present in the database.
Conclusion
Using a smaller database with restriction is important to make sure you see all matching sequences. To find all homologs including paralogs requires the more sensitive blastn or a protein level search.
Always search the smallest database that is likely to contain the matches of interest.
Saved ResultsOn your own exercise:
Repeat this search again using blastn. Do you find additional creatine kinase genes in whales?
Using experimental organism-based nt
The nr/nt database is very large — currently 97 million sequences; 1.3 trillion (1.3 X 10^12) total bases. As it continues to grow we will need to produce subsets to maintain efficiency. As we mentioned, one way is to split nr/nt into subsets by organism. Invertebrate animals and bacteria use arginine kinases and phosphoarginine as a way of regenerating ATP. These products are homologs of the vertebrate creatine kinase. We can use a blastn search to find some bacterial arginine kinases using the new Prokaryote split nt.
Goal
Try out the new experimental taxonomic nt databases and use blastn to identify some bacterial arginine kinases. Understand the differences between the megablast and blastn BLAST programs.
Search setup
Use the same Query sequence and Search type as in the previous examples.
Database, Limits, and Filters
Select the Experimental databases radio button and choose the Prokaryota (bacteria and archaea) nt (nt_prok) database
BLAST program
The parameters for megablast are tuned to find nearly identical matches. The blastn program uses parameters that make it slower but more sensitive. In fact, megablast would find no significant matches for this search.
Change the program to blastn and notice how the parameters change
Run it!
Results
Descriptions
The Descriptions show eight matching database sequences. Notice that with blastn we are more likely to generate alignments that are near the Expect cut off (0.05). The last match has an Expect value of 0.02.
These are all matches to submitted complete genome sequences for bacteria. Some of these have corresponding RefSeq versions that are not in the nr/nt database. You can search the RefSeq Representative genomes database (limited to bacteria) to find RefSeq versions of some of these.
Graphic Summary
The Graphic Summary shows that the all matches are to the central region of the query sequence. At the protein level, this corresponds to the best conserved regions of the protein conserved domain
Alignments
Try changing the 'Alignment view' options to 'Pairwise with dots for identities' and adding the CDS feature to better see conservation at the nucleotide and protein translation level.
Follow the 'Graphics' link for the alignment for Oceanithermus profundus DSM 14977, complete genome to see the match in context on the genome sequence. Notice the Genes track that shows the match to the coding region of an arginine kinase.
Conclusion
By using the smaller prokaryote subset of nr/nt and the more sensitive blastn algorithm you were able to find arginine kinase genes in bacteria
Keep in mind that this type of search should be done at the protein level as proteins are better conserved providing a longer evolutionary look-back time. A way to do that with a nucleotide database is to use the tblastn program that translates the nucleotide database on the fly and allows you to search with a protein query. This isn't yet implemented for the experimental nr/nt subsets. But we'll use tblastn with a genome assembly database as a demo in a few minutes.
Saved Prokaryotic nt Results
Annotating creatine kinase genes in the grey whale assembly
Goal
To annotate creatine kinase genes in the unannotated grey whale assembly using nucleotide BLAST
Search setup
Use the Genomes search box on the BLAST homepage to search for 'Grey whale'. This will take you to the Grey whale genome BLAST page.
Query sequence
Use the human muscle creatine kinase transcript (NM_001824.5) as in the previous examples.
Database, Limits, and Filters
The database is preset on this page to the nucleotide sequences for Grey Whale genome assembly (mEscRob2.pri GenBank assembly [GCA_028021215.1]) that contains assembled chromosome sequences and component sequences.
BLAST program
Run the search the first time with megablast. You can run it again with blastn to see any improvements in the results.
Run megablast!
Megablast results
Descriptions
There are two database sequences with matches, the assembled chromosome 19 sequence, CM051683.1, and the underlying contig sequence. Focus on the matches to assembled chromosome 19, CM051683.1. The Max Score and the Expect value reported here are for the single best match, but there are several matches. The sum of the scores for all matches are given in the Total score column. These matches are to some of the exons of the gene. Notice that the query coverage is less than 100 percent indicating part of the query doesn't align
Graphic summary
Take a look at the graphic summary and note the black vertical lines. As you saw earlier these essentially separate the exon matches and represent regions of the database sequence that don't align. There is also region of the query centered around 600 bases that doesn't match under these conditions. This is because of the lower sensitivity of the megablast program.
Alignments
In the Alignments, you can sort the six matches to to chromosome 19 by 'Query start position' to get them in exon order and add the coding sequence from the annotation of the human transcript.
Set up blastn search
Use 'Edit Search' to try the search again and set the BLAST program to blastn.
Run blastn!
blastn results
Descriptions
There are now matches from four different assembled chromosomes: 19, chromosome 1, chromosome 2, and chromosome 10. The match to chromosome 10 may not be interesting as it's close to the Expect cut-off. The results for chromosome 19 now show 100% query coverage, which indicates that you have all exons.
Graphic summary
Use graphic summary to verify the 100% query coverage for the match to chromosome 19. The matches to chromosome 1 also show some of the exon structure.
Alignments
Examine the alignments for chromosome 1. Follow the 'Graphics link' at the head of the alignments to view the matches in the context of the chromosome. There are two regions that align at different parts of the chromosome. You can zoom in to see that these represent distinct sets of exons probably for different creatine kinase genes. See the image below
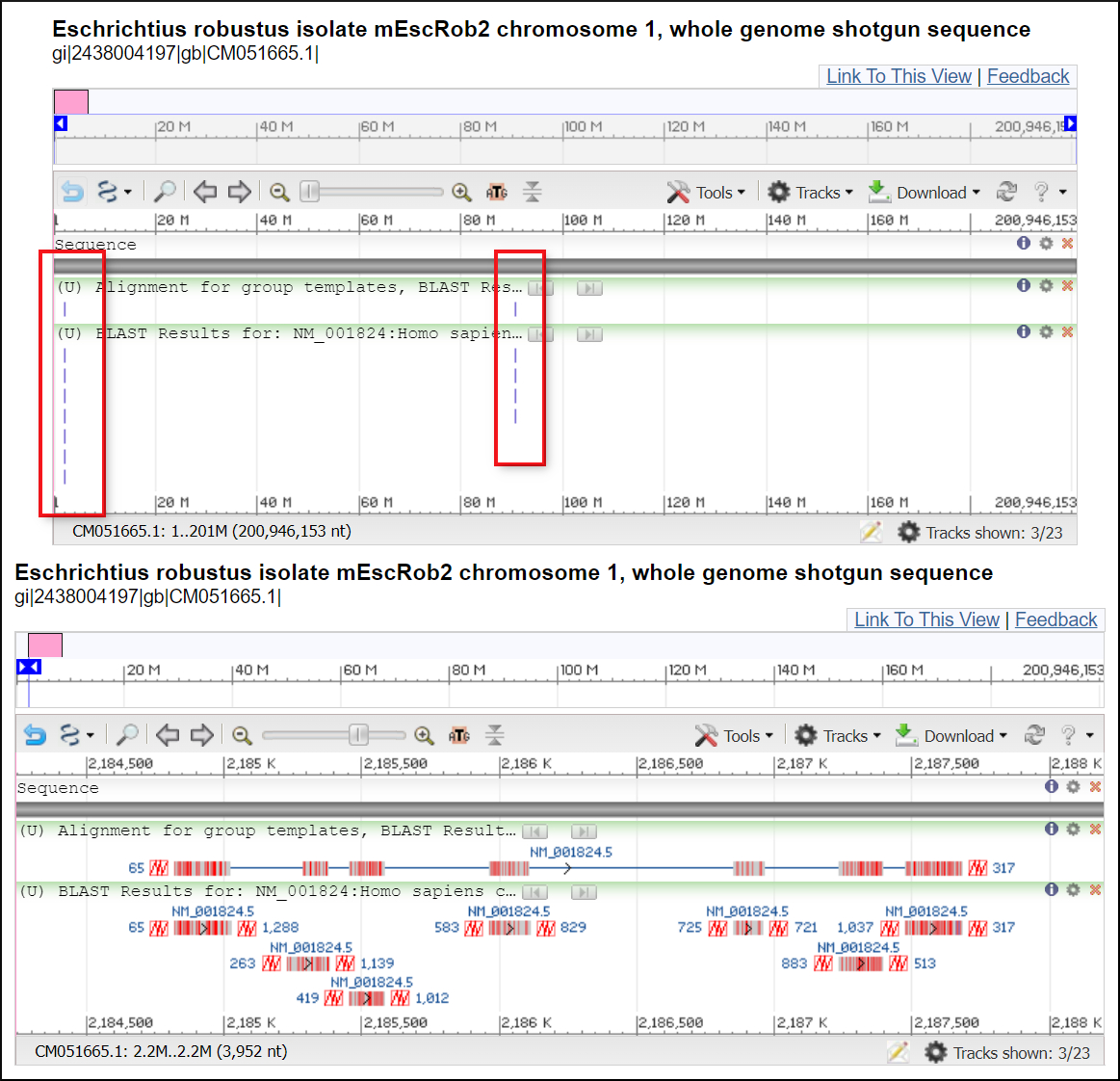
Interpretation and Conclusion
Megablast quickly identifies the location of the muscle type creatine kinase gene on chromosome 19 in the grey whale using a human transcript as a query. The more sensitive blastn finds the location of all exons of the muscle creatine kinase ortholog and is needed to find other paralogs in the genome using the human transcript with the default settings. The blastn search finds the location of four potential creatine kinase orthologs. Using a CKM transcript sequence from another whale would probably give more complete results that with the human transcript.
Demo: Using a translating search (tblastn) to find CKM paralogs in the grey whale
tblastn is a protein level search that generates a protein database of the six-frame translation of the chosen nucleotide database. You can use the human muscle creatine kinase protein to find protein level matches to in the grey whale genome. The cross-species search at the protein level should be more sensitive than the megablast or blastn searches.
Goal
Use a tblastn search to find all creatine kinase homologs in the Grey whale genome
Search setup
Query sequence
Use the human RefSeq Select muscle creatine kinase protein (NP_001815.2)
Search type
You can click 'Edit Search' and change the Search type to a tblastn search. Select the tblastn tab on the query box .
Run tblastn!
Results
Descriptions
We have matches to chromosome 19, 1, and 2 as before. Notice that the chromosome 10 match is not found. One of the chromosome 1 exons is now a best match. However it has a lower percent identity than the chromosome 19 match. The chromosome 1 'Total score' is the highest because there are two genes as we have seen.
Alignments
The alignments are protein alignments. The subject coordinate system is that of the assembled chromosome. The sequence shown is translation frame that matches. Sort by query start position on the first subject to see that there are the two distinct matches on chromosome 1. The other two chromosomes have one region each.
Interpretation and conclusion
Using a protein level search we can identify at least four different creatine kinase genes in the unannotated grey whale assembly. This compares favorably to the four genes annotated by NCBI in the blue whale and minke whale, two other baleen whales.

