ClusteredNR: blastx and PSI-BLAST
Like the default nucleotide database, the protein nr database is also large — 595 million sequences and 234 billion residues — and growing rapidly. In addition, certain kinds of proteins and groups of organisms such as mammals and their associated bacteria are over-represented and will dominate protein searches. This makes it difficult to identify more distant homologs in BLAST searches.
One strategy to collapse some of the over-representation is to form clusters at less than 100% identity.
The experimental ClusteredNR has clusters where the members are 90% identical and within 90% sequence length. This provides access to more distant matches and reduces the computational burden of searching entire nr since BLAST only searches the representative sequence for each cluster. ClusteredNR currently has 286 million sequences and 94 bilion total residues — about 40% the size of the full nr. You'll use this database to expand the results for a blastx search and as a way of incorporating more distant matches in a PSI-BLAST search.
Spiny rat mRNA and ClusteredNR
The Ryukyu spiny rat is endemic to Amami Ōshima island near Japan. It's unusual in that males don't have a Y chromosome. We have a large set of assembled transcript sequences for this species that lack protein annotation. We can use blastx and to annotate a transcript with a protein name and then use the new ClusteredNR database to identify more distant homologs.
Query sequence
Retrieve GHEE01458953.1 and send to BLAST using the 'Run BLAST' link on the right side.
Search type
Select the blastx tab to run a translating search.
blastx searches the a protein database with the six-frame translation of the query sequence.
Database, limits, and filters
Use the default protein nr database for the first search.
You'll use the experimental ClusteredNR database for a second search.
BLAST program
No options for blastx
Algorithm parameters
Use the default settings.
Run blastx against nr!
Results
Descriptions
The Descriptions show 100 matches to neural cell adhesion molecule proteins. There are no exact protein matches as this protein sequence is not in the database. Notice that the query coverage is only 34% for all database matches. This is because the matches are to the coding region on the query and it apparently has long untranslated portions. All matches are to NCBI RefSeq proteins, largely models. The largest Expect value is still rounded to zero, so you don't have all results
Graphic Summary
The graphic shows the coding region match to the query, which apparently has a very long 3' untranslated region.
Alignments
The alignment show the matching reading frame from the query to the database protein sequence.
Taxonomy
All matches all from the rodent superfamily Muroidea (mice, rats, gerbils, hamsters, etc.). You can't see homologs from other groups in this output.
Now, re-run the search against ClusteredNR. You can set the number of descriptions to 500 if you want.
Setup ClusteredNR blastx search
Click 'Edit Search', choose the 'Experimental databases' and select ClusteredNR as the database. In the Algorithm parameters, set the 'Max target sequences' to 500.
Run blastx against ClusteredNR!
blastx ClusteredNR results
Descriptions (Clusters)
The Descriptions show the Clusters instead of the individual matches. The search was performed against a database of the cluster representative sequence. The BLAST statistics (score, evalue, etc.) alignment and other displays are for the representative.
Many clusters are fairly large and collapse hundreds of entries and many organisms. The cluster ancestor is shown in the table. This is the BLAST name of the most most proximal taxon — examples: carnivores, placentals, birds — that contains all organisms with sequences in the cluster
You can access the contents of the cluster by clicking the plus sign (+) for the cluster composition in the left-hand column. To see an example, open a large cluster in the output such as the one for NP_001358111.1 with 297 members.
Each cluster has its own Taxonomy report, and you can download the cluster report and members in many different formats for further analysis. You can also run a BLAST alignment of all the members and then run COBALT if desired.
Taxonomy
The Taxonomy report, which is based on the cluster representatives, shows that this protein has homologs in all jawed vertebrates (Gnathostomata).
Interpretation and Conclusion
The ClusteredNR database offers a good alternative to the protein nr for finding homologs in other species especially when working with query sequences from over represented groups such as rodents.
Saved blastx ClusteredNR results
Using PSI-BLAST with ClusteredNR and a plant defensin
Position Specific Iterative BLAST or PSI-BLAST constructs a position specific score matrix (PSSM) from alignments found in an initial protein BLAST search. You can then search the database again using the PSSM. This second round may collect new significant matches that can be used to add to and refine the PSSM. You can continue over many rounds until the search finds no new significant matches. Researchers use PSI-BLAST to explore deep evolutionary relationships of protein families and conserved domains. PSI-BLAST is the tool used initially to construct the PSSMs in NCBI's Conserved Domain Database.
Because of the increased size of the protein database and the fact that certain groups of organisms are over represented, it's usually very difficult to collect a complete set of protein matches in the first round of PSI-BLAST and continue on to iterate the search. The new ClusteredNR database offers a reduced protein database that can make it possible to run a PSI-BLAST search without requesting huge numbers of matches.
Goal
Identify more distant homologs of a plant protein using PSI-BLAST
Search setup
Query sequence
Retrieve accession AES68994.1, a barrel medic defensin-like protein, from the protein database and send it to BLAST using the 'Run BLAST' link on the page.
Search type
Keep the default protein search (blastp).
Database, limits, and filters
Use the default protein nr database for the first search.
You'll use the experimental ClusteredNR database for a second search.
BLAST program
Choose PSI-BLAST.
Run PSI-BLAST against nr!
PSI-BLAST nr results
Descriptions
The PSI-BLAST results against nr show matches only up to an expect value of 0.001 with the default 500 target sequences. This expect value is lower than the default inclusion threshold for PSI-BLAST (expect value 0.005). PSI-BLAST works by generating a position specific score matrix (PSSM) from the information in the BLAST alignments below the threshold. Since the inclusion threshold wasn’t reached, the results from nr are missing information from more distant matches that may be important to include in the PSSM. Also, a useful feature of web PSI-BLAST is that you can manually select matches above threshold to add their alignment information to the PSI-BLAST PSSM if desired. However, here you don’t have access to any matches above threshold.
In a case like this where you don’t reach the inclusion threshold, you may want to edit the search to get more than the default 500 matches and run it again so you can include all relevant proteins and be able to select matches above threshold.
Setup ClusteredNR PSI-BLAST search
Click 'Edit Search', choose the 'Experimental databases' and select ClusteredNR as the database.
Run PSI-BLAST against ClusteredNR!
PSI-BLAST ClusteredNR results
Descriptions
Using ClusteredNR the results now allows you to reach the PSI-BLAST threshold and see results that are below threshold. You could select which results are included in the PSSM that is used in the next iteration.
Running PSI-BLAST iterations
Click the button on the PSI-BLAST results to run PSI-BLAST iteration 2
PSI-BLAST now searches the ClusteredNR database using a PSSM generated from the sequences in the first round. This will find additional sequences with this more sensitive search method
As shown below, the results will now highlight new sequences in the Descriptions section that are now significant matches when the PSSM is used to search the database in iteration 2.
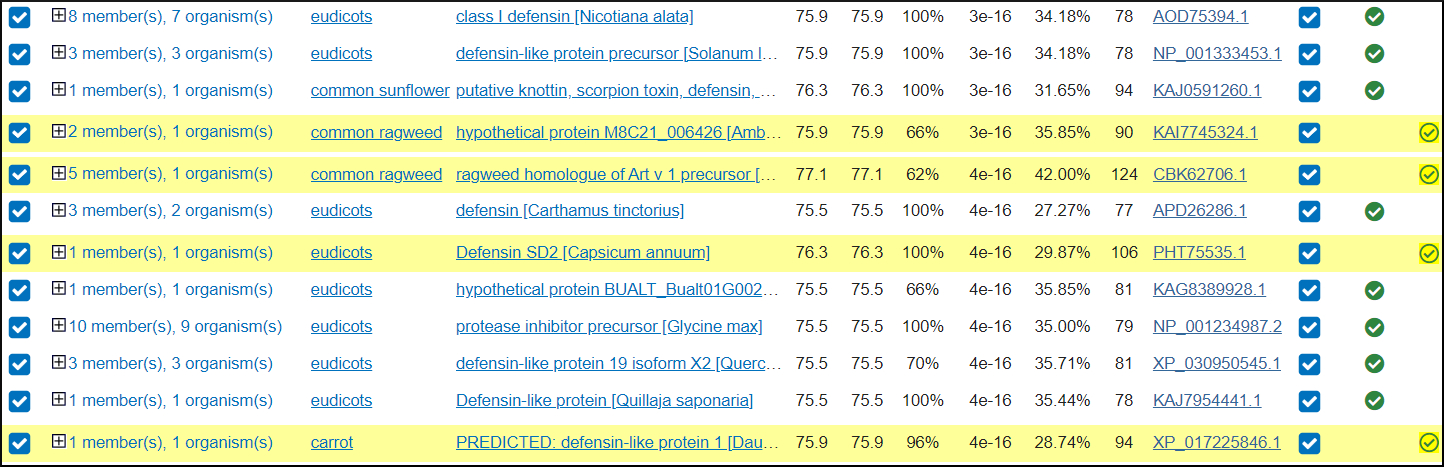
Saved PSI-BLAST ClusteredNR results
Last Reviewed: July 10, 2023

