BLAST Quick Start
This is a quick introduction to running a BLAST search. Each example today will have the same structure as this one.
-
Goal
-
Search set up
-
Query sequence
-
Search type
-
Database, Limits and Filters
-
BLAST program
-
Algorithm parameters
-
Running the search
-
Results
-
Conclusion
Goal
For the introductory search your goal will be to identify closest well-characterized mRNA sequence match in mouse to an unannotated transcript from the Ryukyu spiny rat.
Search set up
Query sequence
Use the Ryukyu spiny rat mRNA sequence, GHEE01192317.1
You can retrieve the sequence from the Nucleotide database and send as a query sequence to BLAST though the 'Run BLAST' link.
Or go directly to the BLAST homepage and select the type of search you want to perform.
The submission form accepts NCBI sequence identifiers, or sequences in FASTA or raw format. You can submit multiple sequences in a single search.
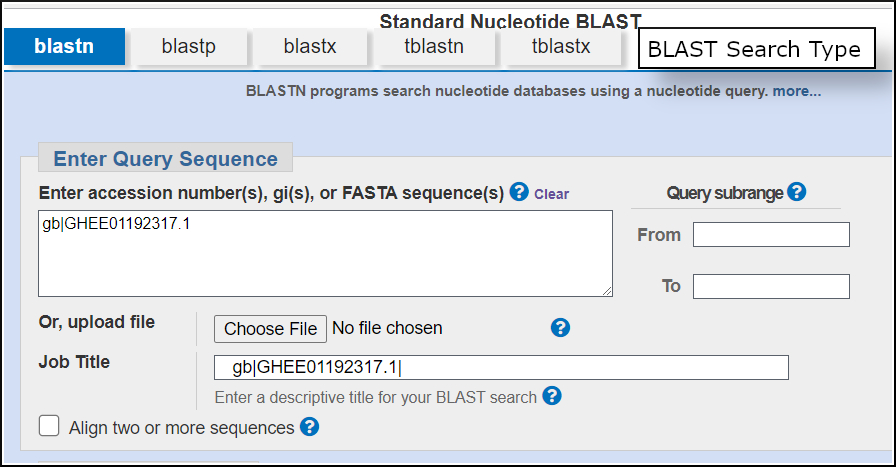
Search type (nucleotide, protein, translations)
You can use the tabs to select the type of search. For a nucleotide sequence like this one, blastn and the query-translating blastx are possible options.
Choose database
We'll use the RefSeq Select database. It contains selected mouse, rat, and human transcript sequences, one transcript per gene. For this purpose it is the easiest database to find a match for an unknown rodent transcript.
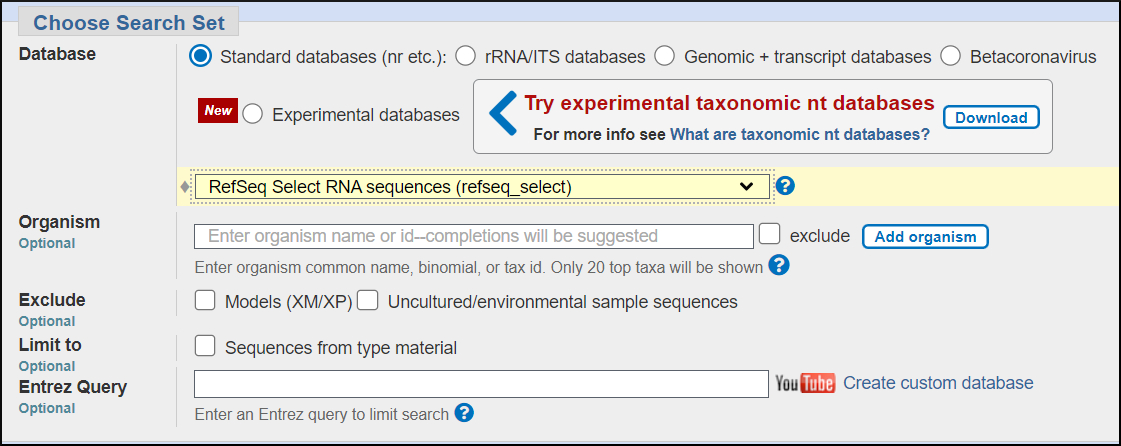
Database modification
In later examples you'll modify the database further to enrich it for sequences of interest and remove those you don't want. You can use the Organism limit and the Sequence type exclusion checkboxes to do this.
Organism limit — restrict to or exclude sequences from NCBI taxonomic groups
Sequence type exclusion — exclude types of records that you don't want in your results such as gene models and uncultured/environmental sample sequences
Select BLAST program
For nucleotide searches the default program is megablast. This is faster but less sensitive than blastn, as we will see in a few minutes.

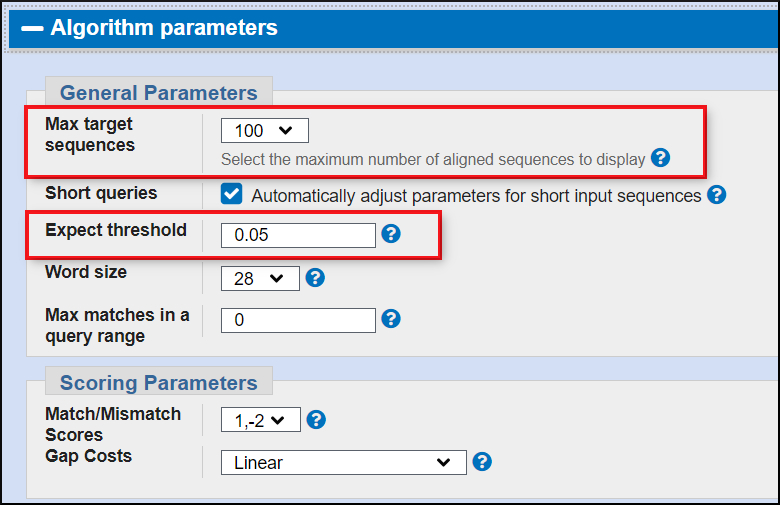
Algorithm parameters
These are more advanced settings for the BLAST programs. Two important ones are Max target seqs and the Expect cutoff.
Max target seqs (default 100) -- Number of matches returned
Expect cutoff (default 0.05) -- Highest Expect value returned. More information on the Expect value below.
Note: Often the Max target sequences setting governs your output. To see all results you may need to increase this setting.
Click the BLAST button to run the search!

Understanding your results
The BLAST output has two main main sections: the Header area and the Tabbed Results.
Header area
This section contains Information about the search (query, database, program)
It also shows the identifier for your search, the Request ID (RID). The RID allows you to access or share your results. It is stable for 36 hours. Send the RID to blast-help@ncbi.nlm.nih.gov if you have trouble with your search.
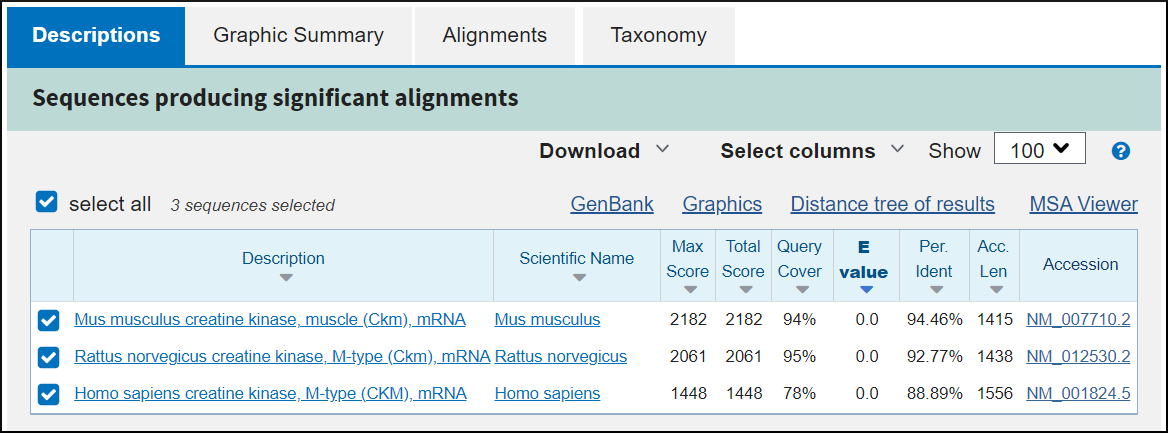
Tabbed Results
Descriptions shows database matches sorted by significance (BLAST Score and Expect value).
BLAST Score — the sum of the match scores (positive), mismatch and gap penalties (negative) in the BLAST aligment.
The nucleotide programs (megablast, blastn) use a simple match / mismatch scoring system with a positive score for an identical match and a negative score for a mismatch and gap open and extend penalties.
The standard protein programs (blastp, blastx, tblastn) use a scoring system based on frequencies of amino acid substitutions in related proteins. The default scoring matrix is BLOSUM62.
Expect Value (e-value) — for a particular match, the number of chance alignments expected with the same score or a better one.
If the e-value is << 1 then the match is not due to chance.
You found significant matches to transcript sequences from mouse, rat, and human. All three are named as transcripts of the creatine kinase m (muscle) gene in each species. The mouse and rat transcripts have better scores than the human one. The e-value of each of these matches have been rounded to zero because the number is so small. There is no doubt that these are non-chance matches.
Graphic Summary
The Graphical Summary shows how the database matches align to the query sequence. All three database matches align to most of the central part of the query sequence.
Alignments
This tab shows details of the local alignment between the query sequence and the database sequence including coordinates on both, gaps, mismatches, and BLAST scores and statistics. The mouse and rat sequence alignments are nearly the same. The human transcript aligns over less of the transcript.
Taxonomy
The Taxonomy tab provides an view of the matches by organism and the organism classification in NCBI's Taxonomy. All hits are to Euarchontoglires, the mammalian group that includes rodents and primates.
There are number of other reports and display options available here that we'll use later on in the workshop.

Interpretation and conclusion
The spiny rat transcript is likely from the homolog of the mammalian muscle creatine kinase gene present in mice, rats, humans and, as we'll see, all other vertebrates.
Homologs — biological features including genes and their products that are descended from a feature present in a common ancestor. There are two kinds of homologs.
Orthologs — genes separated by speciation events
Paralogs — genes separated by gene duplication events
The muscle creatine kinase genes in the three species are orthologs of each other and are paralogs of the other vertebrate creatine kinase gene products (U,B,S) that we'll encounter today.
Last Reviewed: July 31, 2023

