How BLAST works
(Over) simplified algorithm description
- Finds short seed matches (word hits) between a query sequence and a database sequence.
- The word size parameter controls the size of these initial matches and affects speed and sensitivity. The default nucleotide search program, megablast, uses larger word sizes than the traditional blastn program, which is partly why megablast is faster than blastn, but less sensitive
- Intial matches are extended as alignments until the alignment score declines below a certain threshold.
- BLAST then attempts to extend these ungapped alignments by including gaps.
- The gap open and extend penalities affect the size, number of gaps and the lengh of the alignments
- BLAST ranks and filters matches by how unlikely they are, returning — those that wouldn't be expected by chance
- The expect value parameter sets the stringency of this filtering
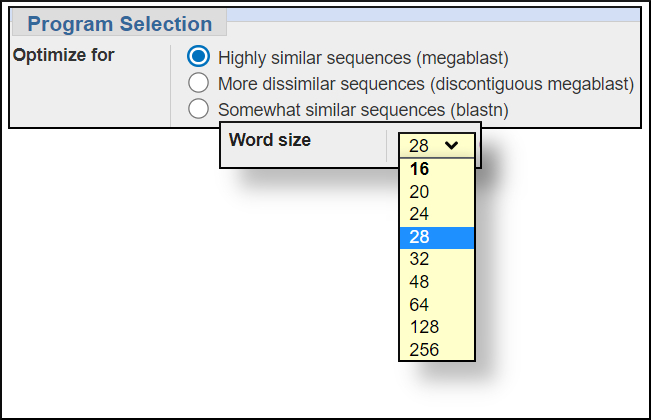
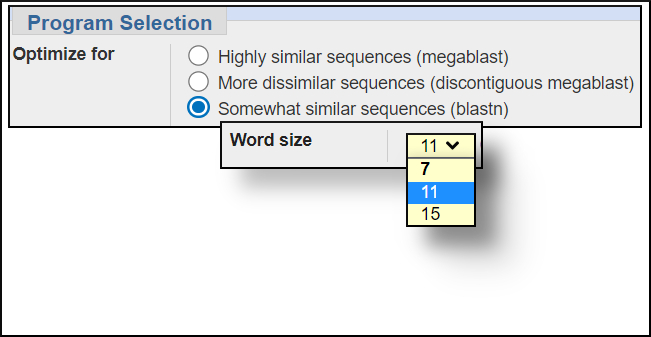
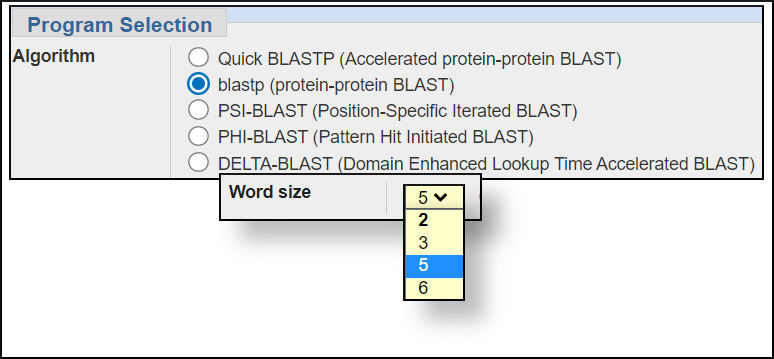
Word size settings
You can change the word size under the Algorithm parameters of the BLAST submission form. In general, larger word sizes increase the speed of a BLAST search and decrease the sensitivity, smaller word sizes make the search run slower but increase the sensitivity.
The available settings for the BLAST programs are shown in the images below. In this workshop we won't change any of the default settings but will see the difference in the results for the same search when using megablast and blastn.megablast word size options
blastn word size options
blastp wordsize options
Last Reviewed: July 2, 2023

